8 Creating Maps using ggplot2
Before you start
In previous chapters, we explored how to quickly create simple maps using vector and raster data. This section will focus on using ggplot2 (along with sf and tidyterra) to produce high-quality maps suitable for publication in journal articles, conference presentations, and professional reports. Achieving this level of quality requires refining the map’s aesthetics beyond default settings. This includes choosing appropriate color schemes, removing extraneous elements, and formatting legends. The focus here is on creating static maps, not the interactive ones commonly found on the web.
Creating maps differs from creating non-spatial figures in some aspects, but the underlying principles and syntax in ggplot2 for both are quite similar. If you already have experience with ggplot2, you will find map-making intuitive and straightforward, even if you have never used it for spatial data before. The primary difference lies in the choice of geom_*() functions. For spatial data visualization, several specialized geom_*() types are available.
-
geom_sf()forsf(vector) objects -
geom_spatraster()forspatraster(raster) objects
These geom_*() functions enable the visualization of both vector and raster data using the consistent and straightforward syntax of ggplot2. They offer direct support for sf and spatraster objects, meaning no prior transformation of these objects is needed before creating maps. In the following sections, 8.1 and 8.2, we will explore each geom individually to understand their basic usage. You will also notice that the principles discussed in the subsequent sections are not specific to spatial data—they are general and applicable to any type of figure.
Note : Although this chapter does not require extensive knowledge of ggplot2, having a basic understanding of it will be extremely helpful. If you are unfamiliar with ggplot2 or feel that your knowledge is insufficient, Appendix B offers a minimal introduction to data visualization using the ggplot2 package. This should provide enough background to help you follow along with the content in this chapter.
Useful resources
As mentioned earlier, a general understanding of how ggplot2 works is highly beneficial. Therefore, any resources for learning ggplot2 can be valuable. Some useful resources include:
The following book offers numerous examples of map-making with ggplot2 and is an excellent resource for further improving your map-making skills after completing this chapter.
Direction for replication
Datasets
All the datasets that you need to import are available here. In this chapter, file paths are set relative to my working directory (which is not shown). To run the code without having to adjust the file paths, follow these steps:
- set a folder (any folder) as the working directory using
setwd() - create a folder called “Data” inside the folder designated as the working directory (if you have created a “Data” folder previously, skip this step)
- download the pertinent datasets from here
- place all the files in the downloaded folder in the “Data” folder
Packages
- Run the following code to install or load (if already installed) the
pacmanpackage, and then install or load (if already installed) the listed package inside thepacman::p_load()function.
if (!require("pacman")) install.packages("pacman")
pacman::p_load(
stars, # spatiotemporal data handling
raster, # raster data handling
terra, # raster data handling
tidyterra, # for tidyverse like operations on terra objects
sf, # vector data handling
dplyr, # data wrangling
stringr, # string manipulation
lubridate, # dates handling
data.table, # data wrangling
patchwork, # arranging figures
tigris, # county border
colorspace, # color scale
viridis, # arranging figures
tidyr, # reshape
ggspatial, # north arrow and scale bar
ggplot2 # make maps
)
8.1 Creating maps from sf objects
This section explains how to create maps from vector data stored as an sf object via geom_sf().
8.1.1 Datasets
The following datasets will be used for illustrations.
Points
-
af_used: total annual groundwater pumping at individual irrigation wells
#--- read in the KS wells data ---#
(
gw_KS_sf <- readRDS("Data/gw_KS_sf.rds")
)Simple feature collection with 56225 features and 3 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -102.0495 ymin: 36.99561 xmax: -94.70746 ymax: 40.00191
Geodetic CRS: NAD83
First 10 features:
well_id year af_used geometry
1 1 2010 67.00000 POINT (-100.4423 37.52046)
2 1 2011 171.00000 POINT (-100.4423 37.52046)
3 3 2010 30.93438 POINT (-100.7118 39.91526)
4 3 2011 12.00000 POINT (-100.7118 39.91526)
5 7 2010 0.00000 POINT (-101.8995 38.78077)
6 7 2011 0.00000 POINT (-101.8995 38.78077)
7 11 2010 154.00000 POINT (-101.7114 39.55035)
8 11 2011 160.00000 POINT (-101.7114 39.55035)
9 12 2010 28.17239 POINT (-95.97031 39.16121)
10 12 2011 89.53479 POINT (-95.97031 39.16121)Polygons
(
KS_county <- tigris::counties(state = "Kansas", cb = TRUE) %>%
st_as_sf() %>%
st_transform(st_crs(gw_KS_sf))
)Simple feature collection with 105 features and 12 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -102.0517 ymin: 36.99302 xmax: -94.58841 ymax: 40.00316
Geodetic CRS: NAD83
First 10 features:
STATEFP COUNTYFP COUNTYNS AFFGEOID GEOID NAME
40 20 175 00485050 0500000US20175 20175 Seward
181 20 027 00484983 0500000US20027 20027 Clay
182 20 171 00485048 0500000US20171 20171 Scott
183 20 047 00484993 0500000US20047 20047 Edwards
373 20 147 00485037 0500000US20147 20147 Phillips
485 20 149 00485038 0500000US20149 20149 Pottawatomie
486 20 055 00485326 0500000US20055 20055 Finney
487 20 167 00485046 0500000US20167 20167 Russell
488 20 135 00485031 0500000US20135 20135 Ness
489 20 093 00485011 0500000US20093 20093 Kearny
NAMELSAD STUSPS STATE_NAME LSAD ALAND AWATER
40 Seward County KS Kansas 06 1656693304 1961444
181 Clay County KS Kansas 06 1671314413 26701337
182 Scott County KS Kansas 06 1858536838 306079
183 Edwards County KS Kansas 06 1610699245 206413
373 Phillips County KS Kansas 06 2294395636 22493383
485 Pottawatomie County KS Kansas 06 2177493041 54149843
486 Finney County KS Kansas 06 3372157854 1716371
487 Russell County KS Kansas 06 2295402858 34126776
488 Ness County KS Kansas 06 2783562234 667491
489 Kearny County KS Kansas 06 2254696661 1133601
geometry
40 MULTIPOLYGON (((-101.0681 3...
181 MULTIPOLYGON (((-97.3707 39...
182 MULTIPOLYGON (((-101.1284 3...
183 MULTIPOLYGON (((-99.56988 3...
373 MULTIPOLYGON (((-99.62821 3...
485 MULTIPOLYGON (((-96.72774 3...
486 MULTIPOLYGON (((-101.103 37...
487 MULTIPOLYGON (((-99.04234 3...
488 MULTIPOLYGON (((-100.2477 3...
489 MULTIPOLYGON (((-101.5419 3...Lines
(
KS_railroads <- st_read(dsn = "Data/", layer = "tl_2015_us_rails") %>%
st_crop(KS_county)
)Simple feature collection with 5804 features and 3 fields
Geometry type: GEOMETRY
Dimension: XY
Bounding box: xmin: -102.0517 ymin: 36.99302 xmax: -94.58841 ymax: 40.00316
Geodetic CRS: NAD83
First 10 features:
LINEARID FULLNAME MTFCC
2124 11051038759 Bnsf RR R1011
2136 11051038771 Bnsf RR R1011
2141 11051038776 Bnsf RR R1011
2186 11051047374 Rock Island RR R1011
2240 11051048071 Burlington Northern Santa Fe RR R1011
2252 11051048083 Burlington Northern Santa Fe RR R1011
2256 11051048088 Burlington Northern Santa Fe RR R1011
2271 11051048170 Chicago Burlington and Quincy RR R1011
2272 11051048171 Chicago Burlington and Quincy RR R1011
2293 11051048193 Chicago Burlington and Quincy RR R1011
geometry
2124 LINESTRING (-94.58841 39.15...
2136 LINESTRING (-94.59017 39.11...
2141 LINESTRING (-94.58841 39.15...
2186 LINESTRING (-94.58893 39.11...
2240 LINESTRING (-94.58841 39.15...
2252 LINESTRING (-94.59017 39.11...
2256 LINESTRING (-94.58841 39.15...
2271 LINESTRING (-94.58862 39.15...
2272 LINESTRING (-94.58883 39.11...
2293 LINESTRING (-94.58871 39.11...
8.1.2 Basic usage of geom_sf()
geom_sf() allows for visualizing sf objects. Conveniently, geom_sf() automatically detects the geometry type of spatial objects stored in sf and draw maps accordingly. For example, the following codes create maps of Kansas wells (points), Kansas counties (polygons), and railroads in Kansas (lines):
As you can see, the different geometry types are handled by a single geom type, geom_sf(). Notice also that neither of the x-axis (longitude) and y-axis (latitude) is provided to geom_sf() in the example codes above. When you create a map, longitude and latitude are always used for x- and y-axis. geom_sf() is smart enough to know the geometry types and draw spatial objects accordingly.
8.1.3 Specifying the aesthetics
There are various aesthetics options you can use. Available aesthetics vary by the type of geometry. This section shows the basics of how to specify the aesthetics of maps. Finer control of aesthetics will be discussed later.
8.1.3.1 Points
- color: color of the points
- fill: available for some shapes (but likely useless)
- shape: shape of the points
- size: size of the points (rarely useful)
For illustration here, let’s focus on the wells in one county so it is easy to detect the differences across various aesthetics configurations.
#--- wells in Stevens County ---#
gw_Stevens <- KS_county %>%
filter(NAME == "Stevens") %>%
st_crop(gw_KS_sf, .)example 1
-
color: dependent on
af_used(the amount of groundwater extraction) - size: constant across the points (bigger than default)
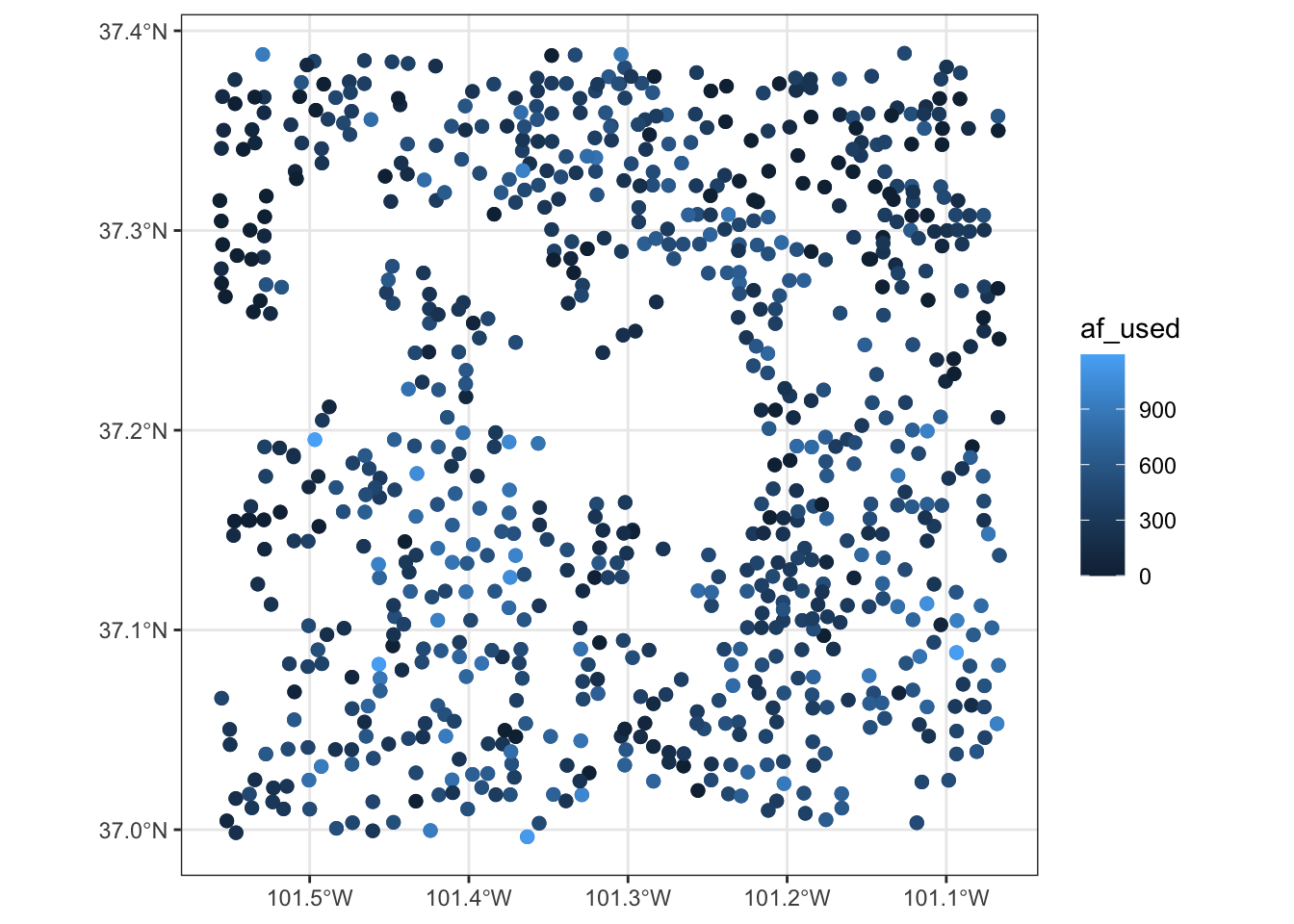
example 2
- color: constant across the points (blue)
-
size: dependent on
af_used - shape: constant across the points (square)
example 3
- color: dependent on whether located east of west of -101.3 in longitude
- shape: dependent on whether located east of west of -101.3 in longitude
(
gw_Stevens %>%
cbind(., st_coordinates(.)) %>%
mutate(east_west = ifelse(X < -101.3, "west", "east")) %>%
ggplot(data = .) +
geom_sf(aes(shape = east_west, color = east_west))
)
8.1.3.2 Polygons
- color: color of the borders of the polygons
- fill: color of the inside of the polygons
- shape: not available
- size: not available
example 1
- color: constant (red)
- fill: constant (dark green)
example 2
- color: default (black)
- fill: dependent on the total amount of pumping in 2010
# In knitting the book, dplyr operations like filter, mutate cause an error even though they would not produce any errors when you are running on an R session. The solution used here is to create the object to be used and save it, and then read it.
# KS_county_with_pumping <-
# gw_KS_sf %>%
# #--- only year == 2010 ---#
# dplyr::filter(year == 2010) %>%
# #--- get total pumping by county ---#
# aggregate(., KS_county, sum, na.rm = TRUE)
# saveRDS(KS_county_with_pumping, "Data/KS_county_with_pumping.rds")
KS_county_with_pumping <- readRDS("Data/KS_county_with_pumping.rds")
KS_county_with_pumping <-
gw_KS_sf %>%
#--- only year == 2010 ---#
dplyr::filter(year == 2010) %>%
#--- get total pumping by county ---#
aggregate(., KS_county, sum, na.rm = TRUE)
ggplot(data = KS_county_with_pumping) +
geom_sf(aes(fill = af_used))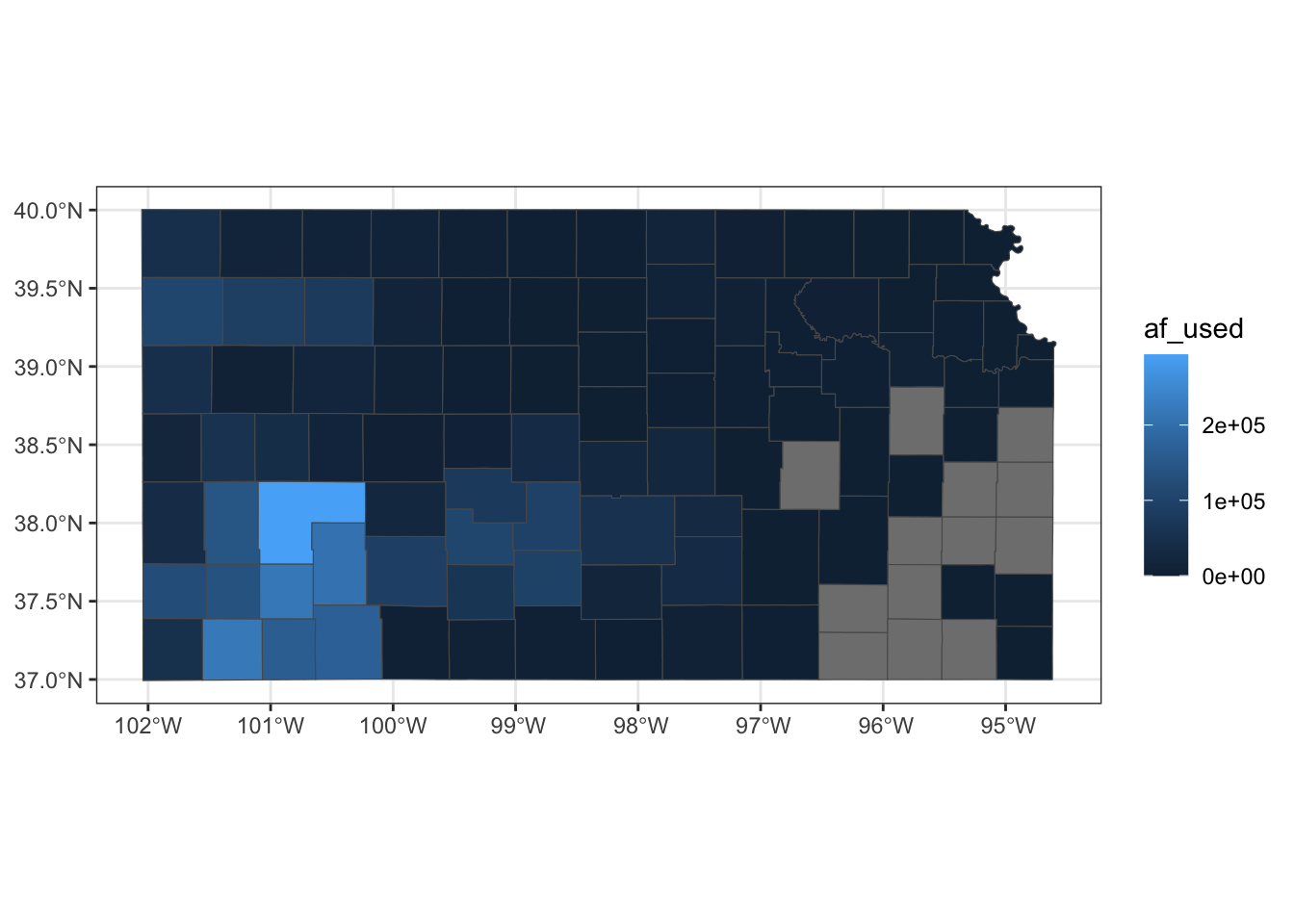
8.1.4 Plotting multiple spatial objects in one figure
You can combine all the layers created by geom_sf() additively so they appear in a single map:
ggplot() +
#--- this one uses KS_wells ---#
geom_sf(data = gw_KS_sf, size = 0.4) +
#--- this one uses KS_county ---#
geom_sf(data = KS_county) +
#--- this one uses KS_railroads ---#
geom_sf(data = KS_railroads, color = "red")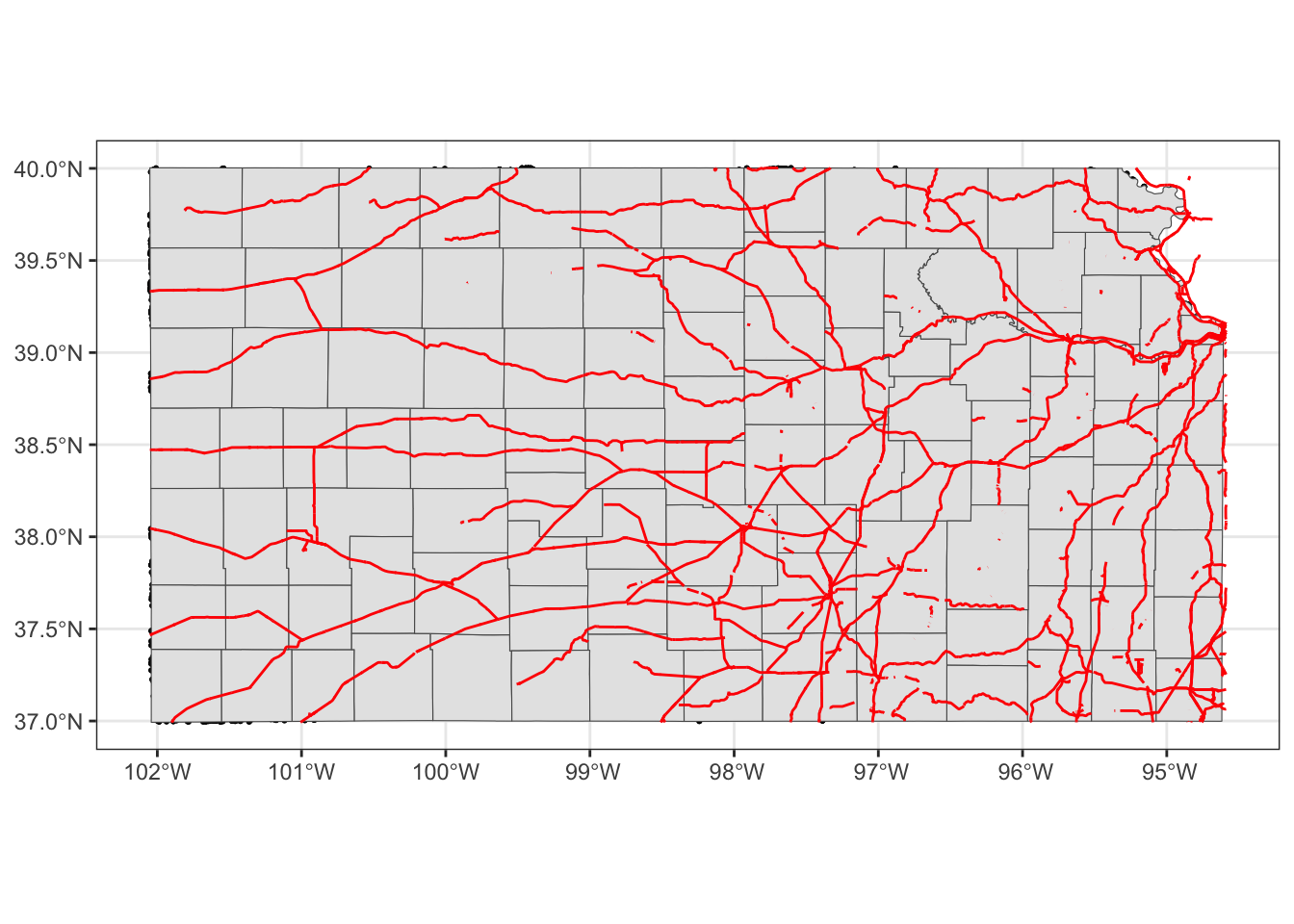
Oops, you cannot see wells (points) in the figure. The order of geom_sf() matters. The layer added later will come on top of the preceding layers. That’s why wells are hidden beneath Kansas counties. So, let’s do this:
ggplot(data = KS_county) +
#--- this one uses KS_county ---#
geom_sf() +
#--- this one uses KS_county ---#
geom_sf(data = gw_KS_sf, size = 0.4) +
#--- this one uses KS_railroads ---#
geom_sf(data = KS_railroads, color = "red")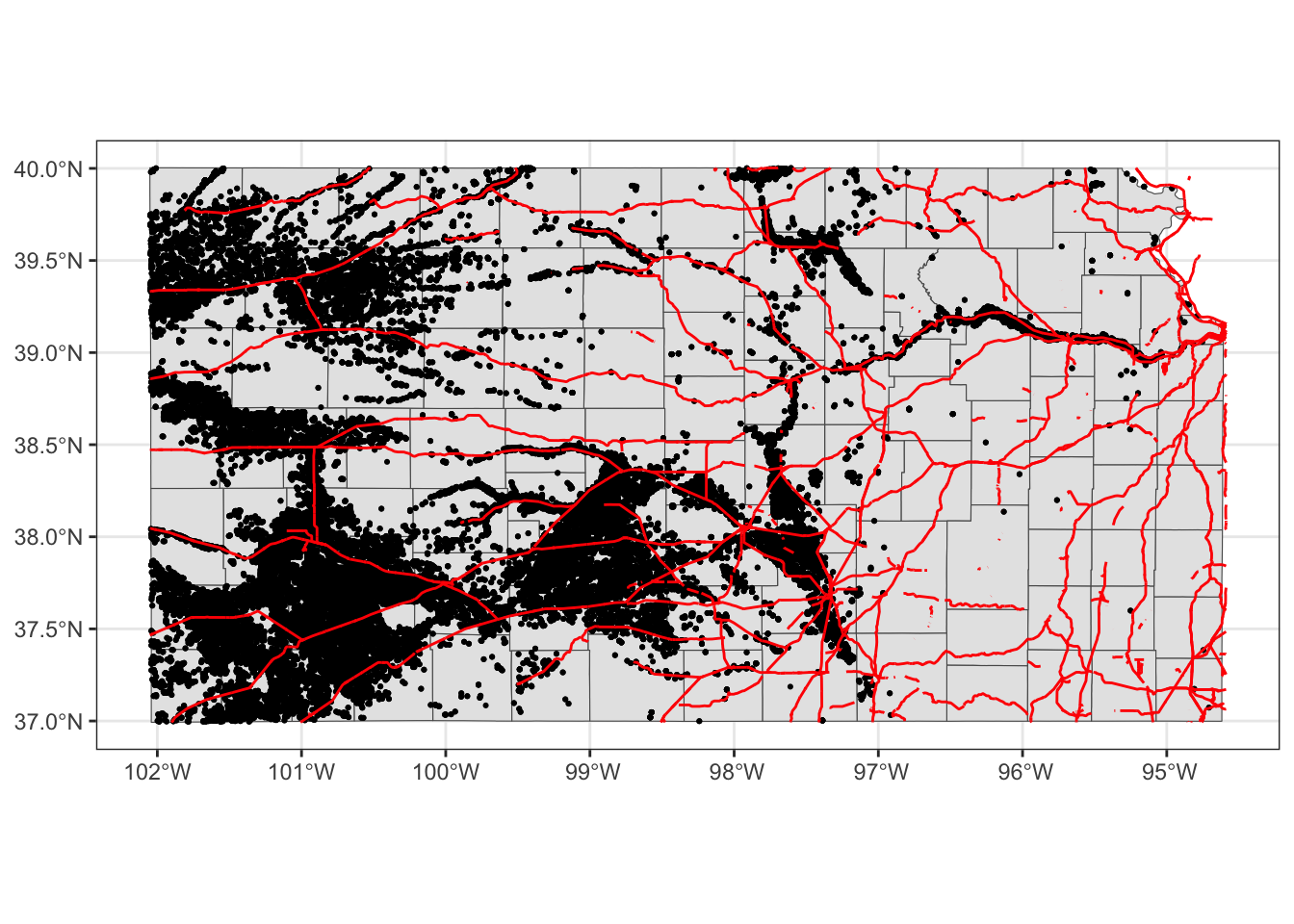
Better.
Note that since you are using different datasets for each layer, you need to specify the dataset to use in each layer except for the first geom_sf() which inherits data = KS_wells from ggplot(data = KS_wells). Of course, this will create exactly the same map:
(
g_all <- ggplot() +
#--- this one uses KS_county ---#
geom_sf(data = KS_county) +
#--- this one uses KS_wells ---#
geom_sf(data = gw_KS_sf, size = 0.4) +
#--- this one uses KS_railroads ---#
geom_sf(data = KS_railroads, color = "red")
)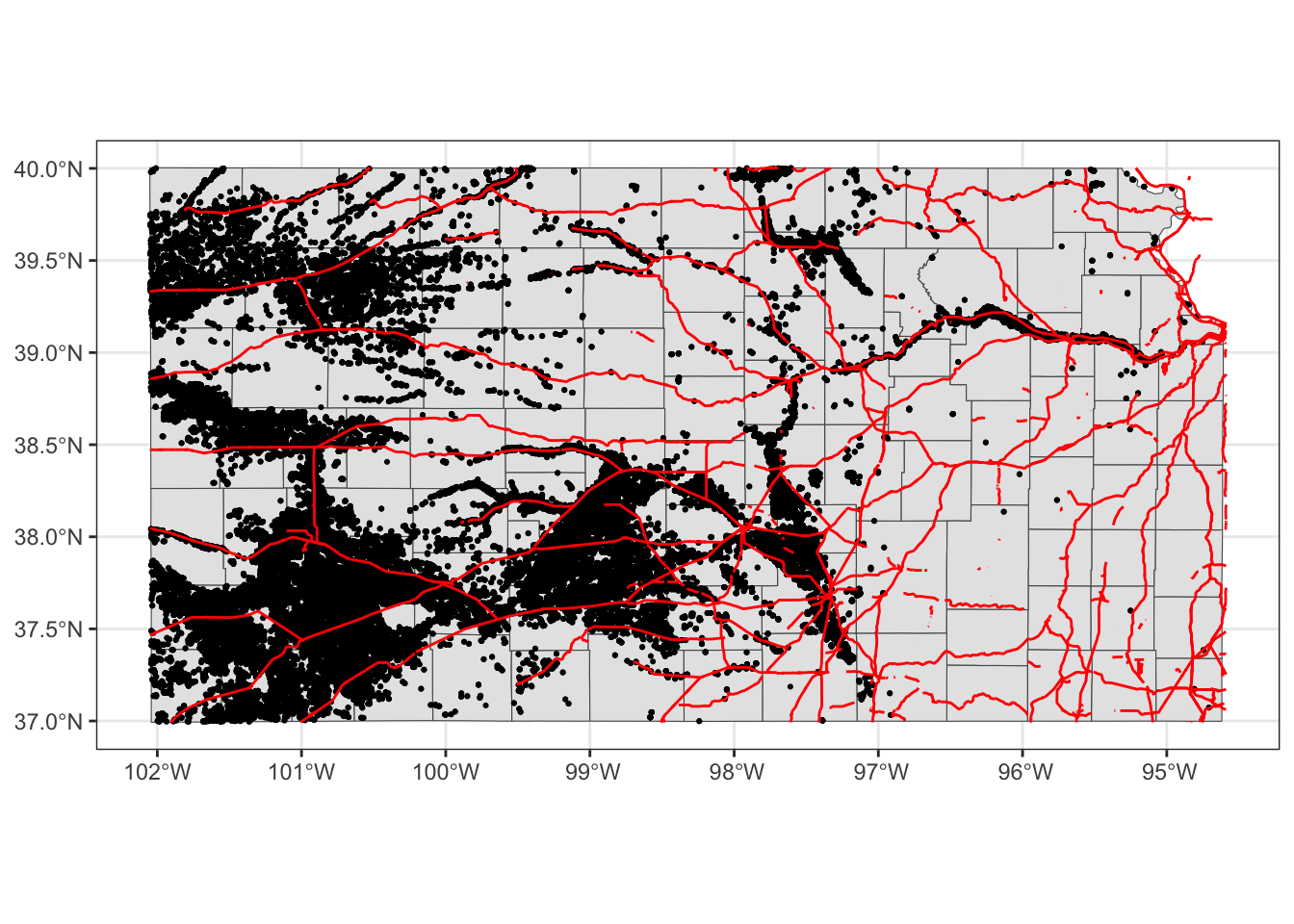
There is no rule that you need to supply data to ggplot().92
Alternatively, you could add fill = NA to geom_sf(data = KS_county) instead of switching the order.
ggplot() +
#--- this one uses KS_wells ---#
geom_sf(data = gw_KS_sf, size = 0.4) +
#--- this one uses KS_county ---#
geom_sf(data = KS_county, fill = NA) +
#--- this one uses KS_railroads ---#
geom_sf(data = KS_railroads, color = "red")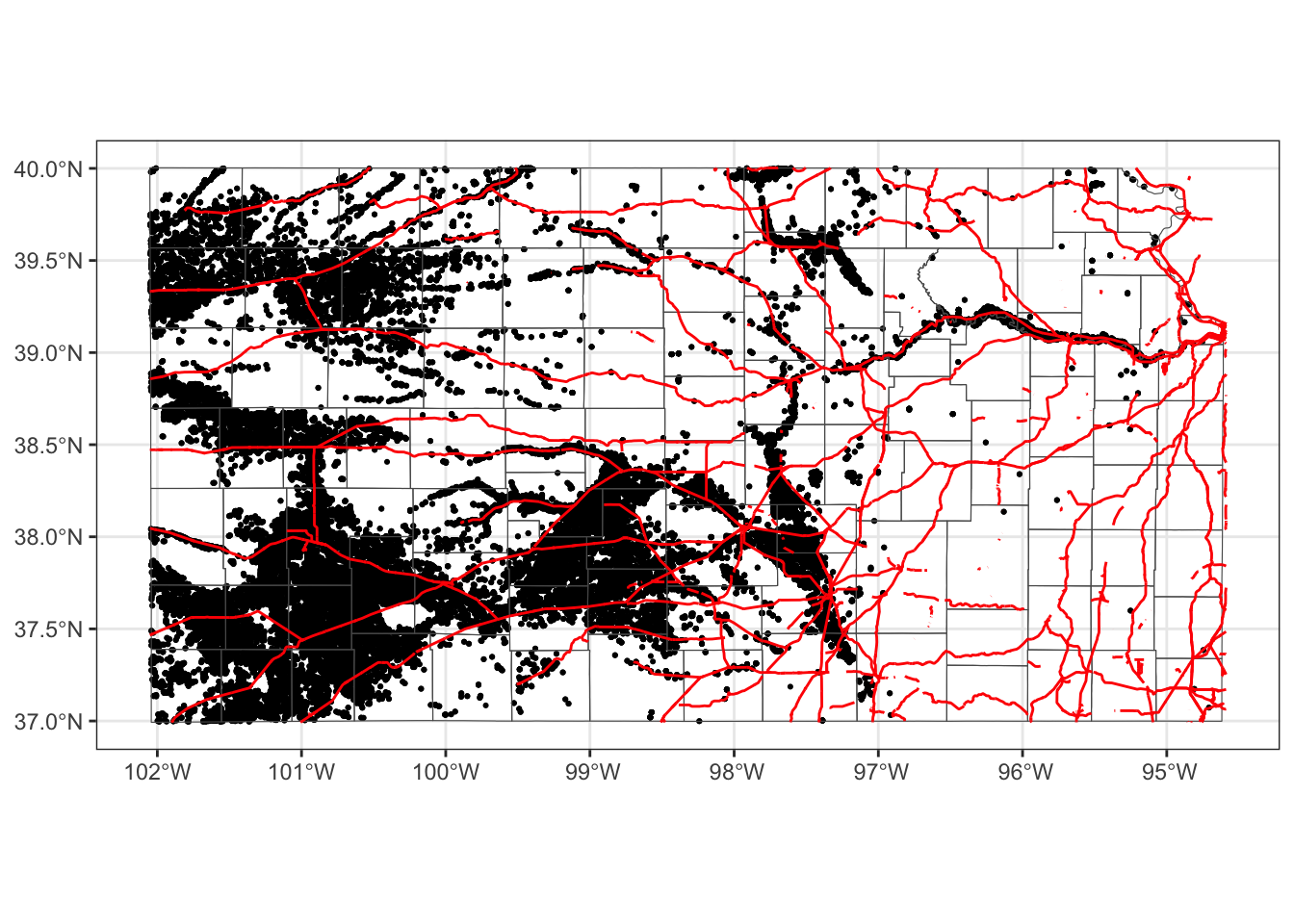
This is fine as long as you do not intend to color-code counties.
8.1.5 CRS
ggplot() uses the CRS of the sf to draw a map. For example, right now the CRS of KS_county is this:
st_crs(KS_county)Coordinate Reference System:
User input: EPSG:4269
wkt:
GEOGCS["NAD83",
DATUM["North_American_Datum_1983",
SPHEROID["GRS 1980",6378137,298.257222101,
AUTHORITY["EPSG","7019"]],
TOWGS84[0,0,0,0,0,0,0],
AUTHORITY["EPSG","6269"]],
PRIMEM["Greenwich",0,
AUTHORITY["EPSG","8901"]],
UNIT["degree",0.0174532925199433,
AUTHORITY["EPSG","9122"]],
AUTHORITY["EPSG","4269"]]Let’s convert the CRS to WGS 84/ UTM zone 14N (EPSG code: 32614), make a map, and compare the ones with different CRS side by side.
g_county / g_32614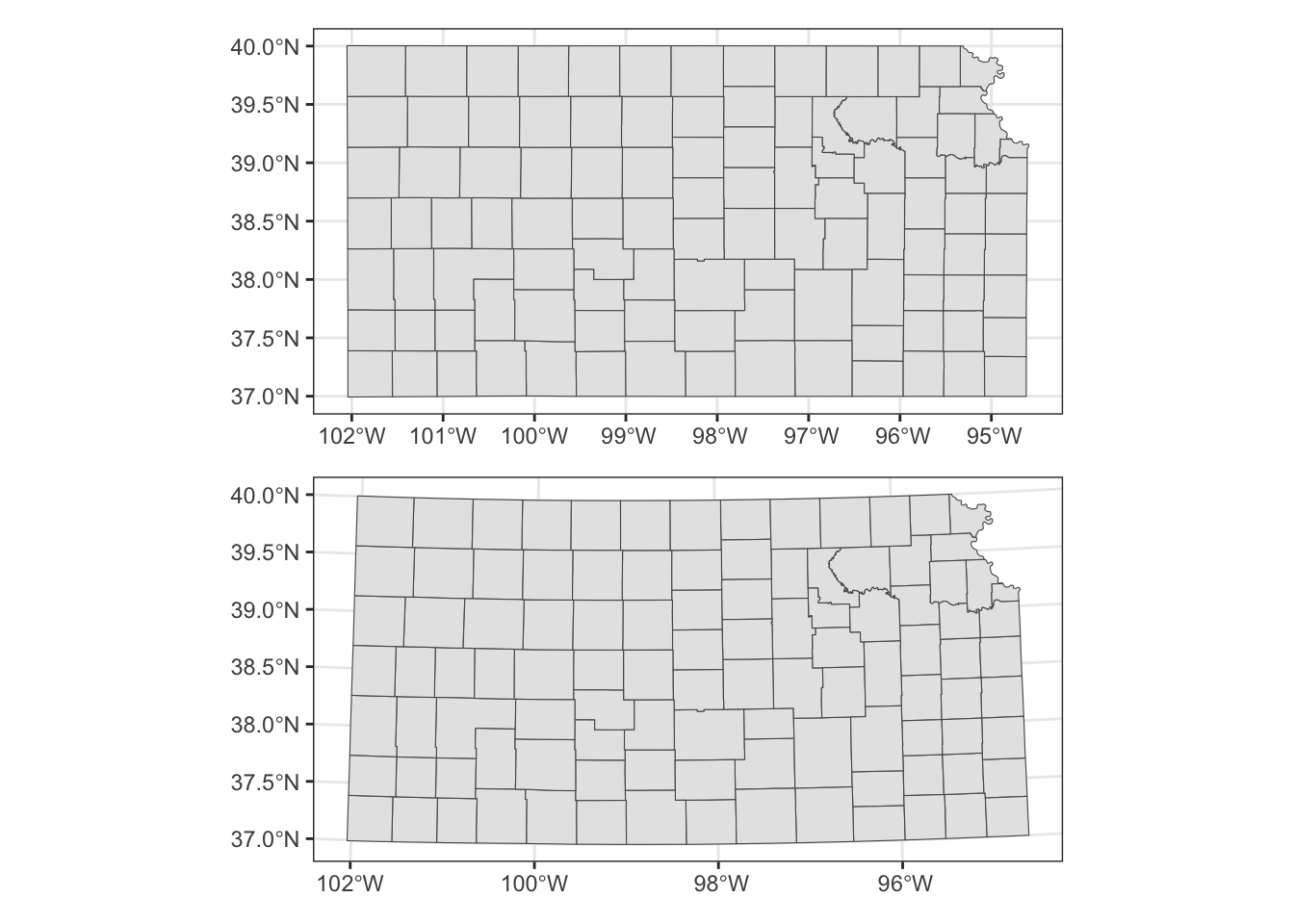
Alternatively, you could use coord_sf() to alter the CRS on the map, but not the CRS of the sf object itself.
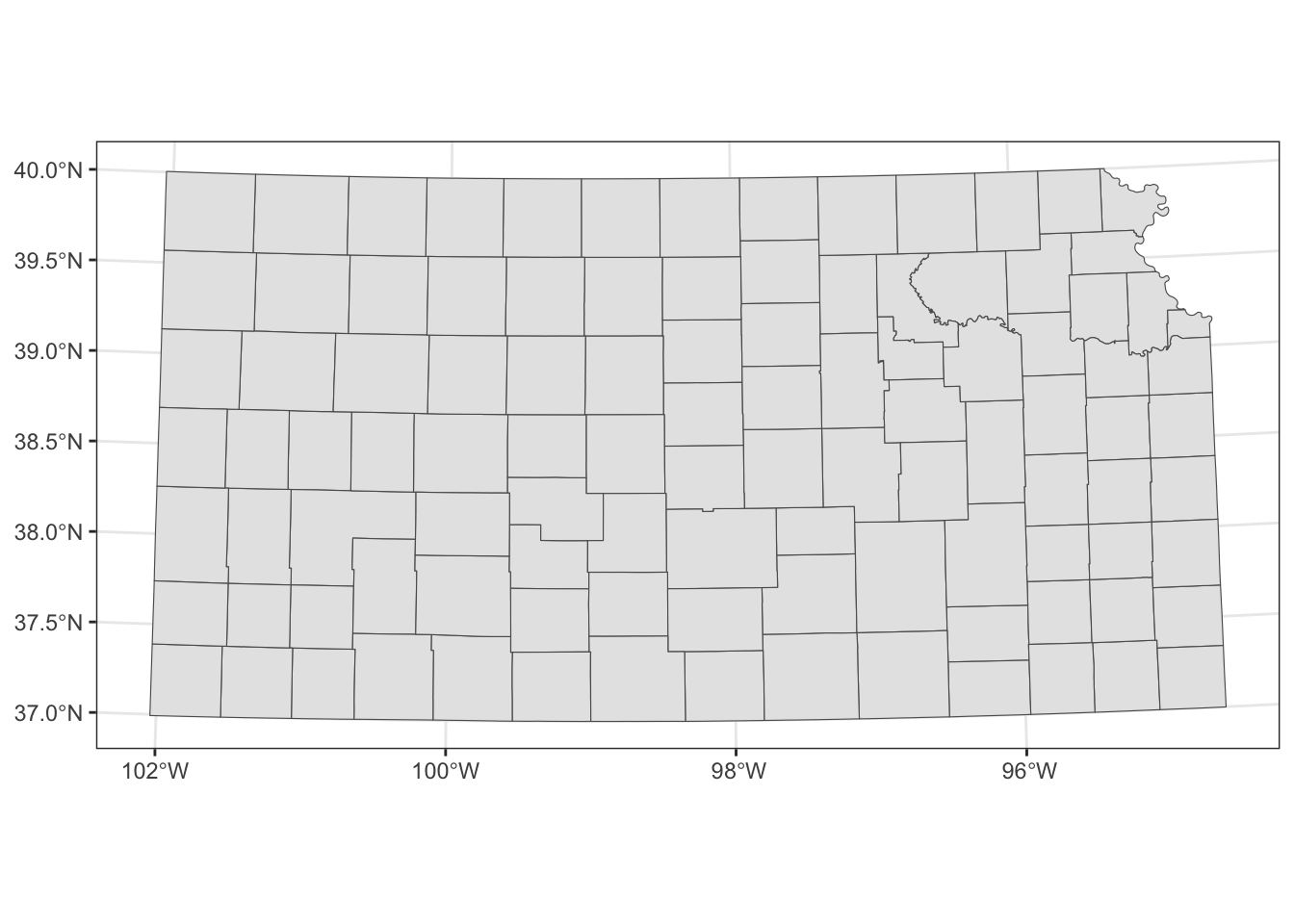
When multiple layers are used for map creation, the CRS of the first layer is applied for all the layers.
ggplot() +
#--- epsg: 32614 ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- epsg: 4269 ---#
geom_sf(data = KS_railroads)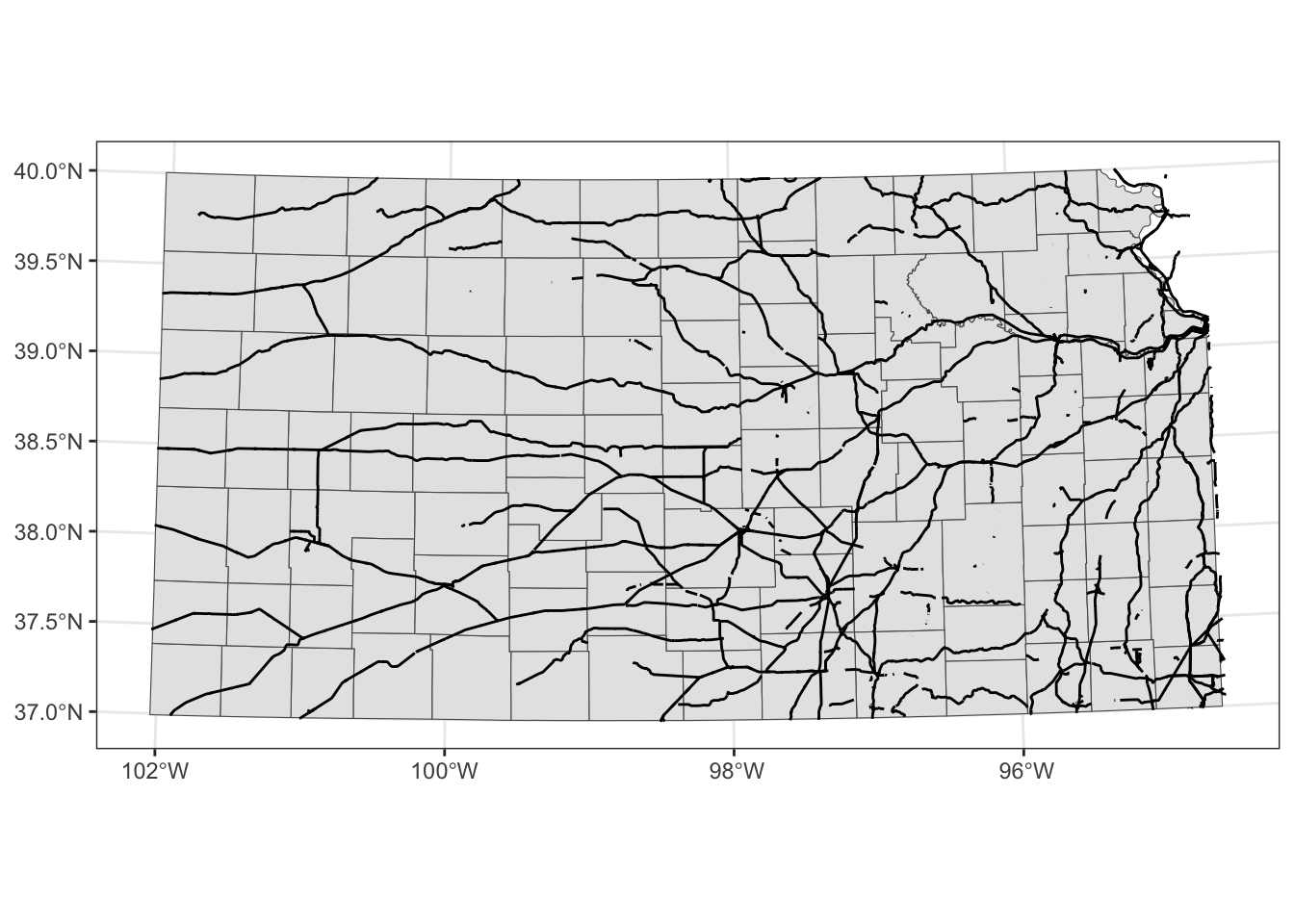
coord_sf() applies to all the layers.
ggplot() +
#--- epsg: 32614 ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- epsg: 4269 ---#
geom_sf(data = KS_railroads) +
#--- using 4269 ---#
coord_sf(crs = 4269)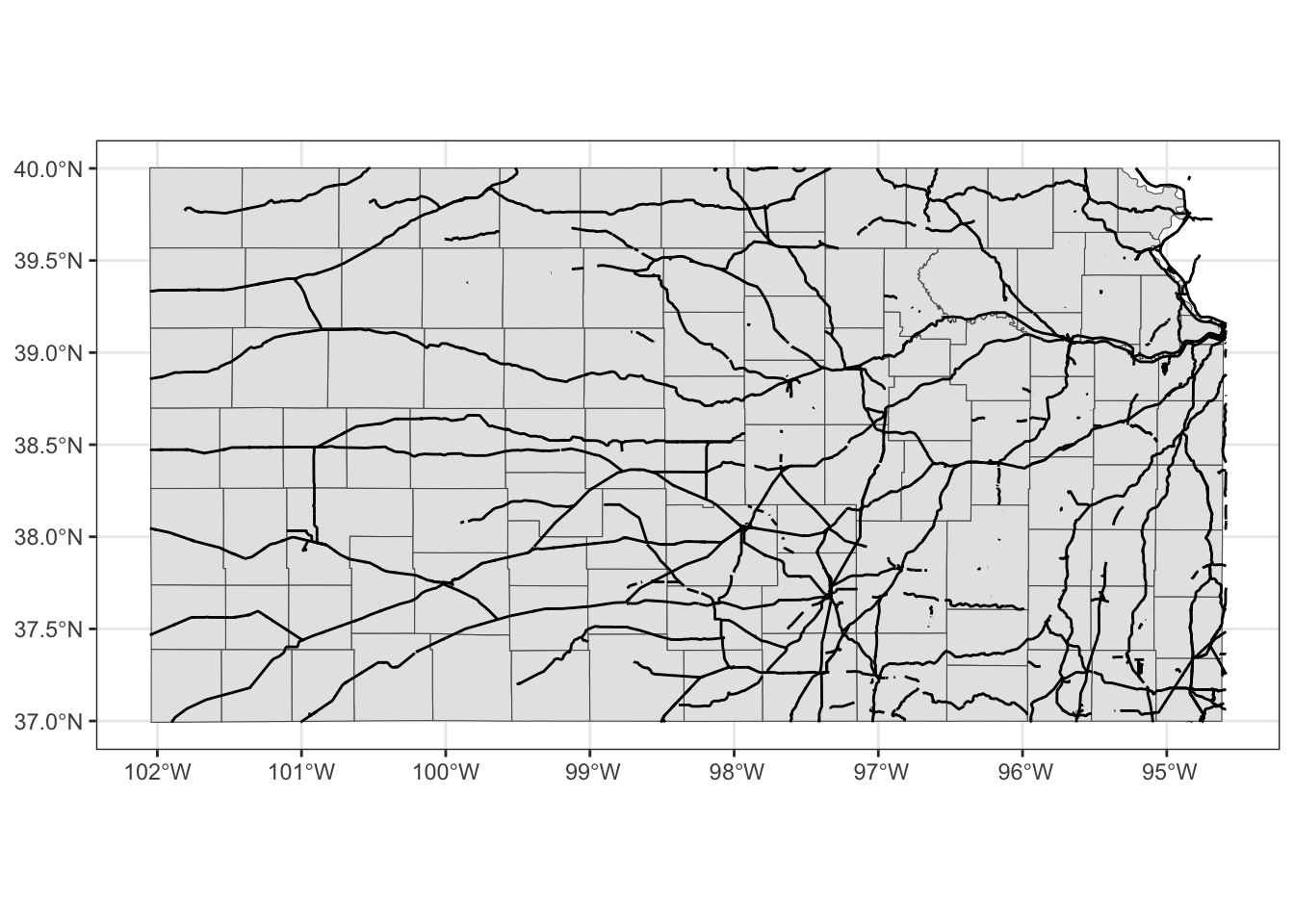
Finally, you could limit the geographic scope of the map to be created by adding xlim() and ylim().
ggplot() +
#--- epsg: 32614 ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- epsg: 4269 ---#
geom_sf(data = KS_railroads) +
#--- using 4269 ---#
coord_sf(crs = 4269) +
#--- limit the geographic scope of the map ---#
xlim(-99, -97) +
ylim(37, 39)
8.1.6 Faceting
Faceting splits the data into groups and generates a figure for each group, where the aesthetics of the figures are consistent across the groups. Faceting can be done using facet_wrap() or facet_grid(). Let’s try to create a map of groundwater use at wells by year where the points are color differentiated by the amount of groundwater use (af_used).
ggplot() +
#--- KS county boundary ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- wells ---#
geom_sf(data = gw_KS_sf, aes(color = af_used)) +
#--- facet by year (side by side) ---#
facet_wrap(. ~ year)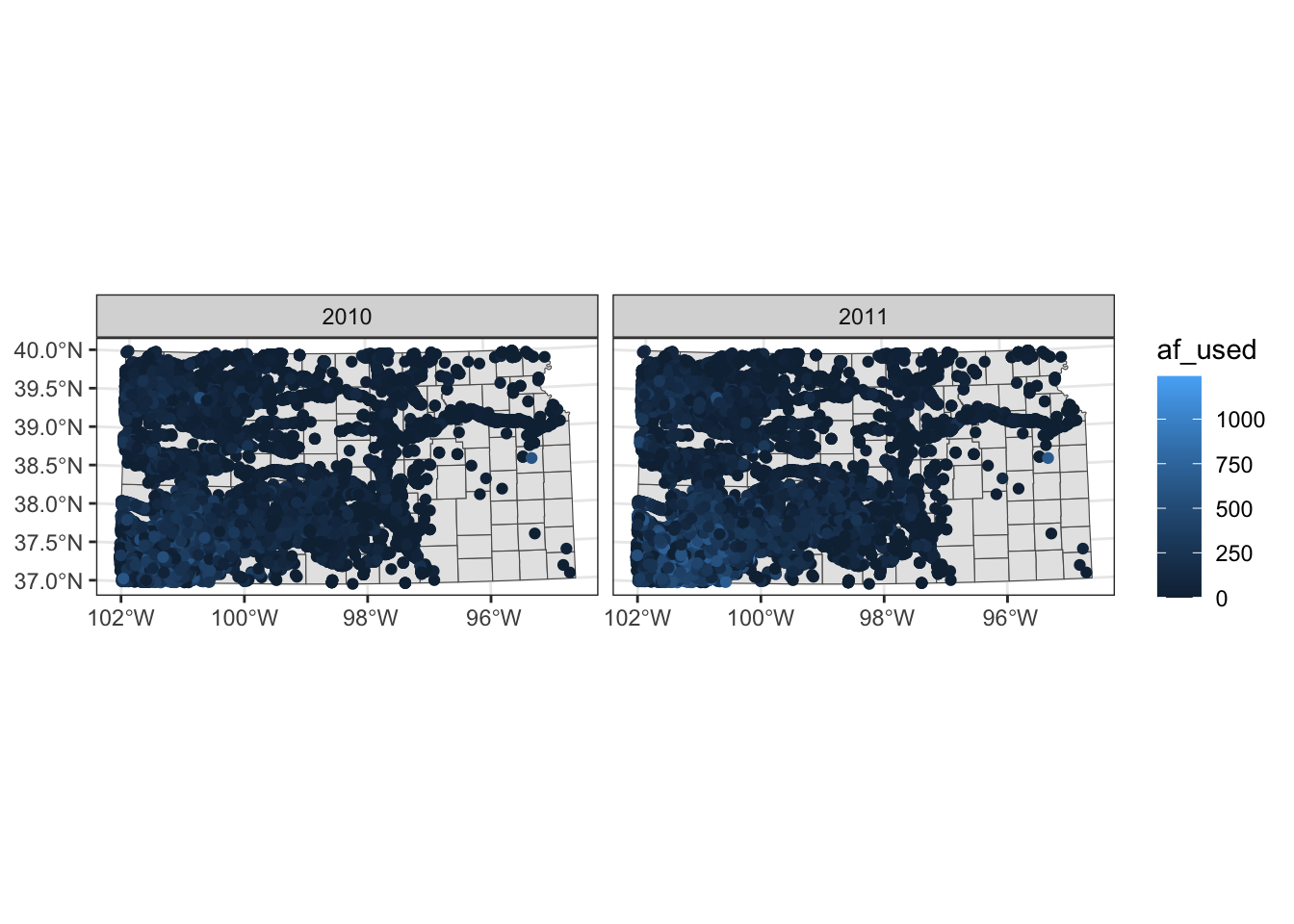
Note that the above code creates a single legend that applies to both panels, which allows you to compare values across panels (years here). Further, also note that the values of the faceting variable (year) are displayed in the gray strips above the maps. You can have panels stacked vertically by using the ncol option (or nrow also works) in facet_wrap(. ~ year) as follows:
ggplot() +
#--- KS county boundary ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- wells ---#
geom_sf(data = gw_KS_sf, aes(color = af_used)) +
#--- facet by year (side by side) ---#
facet_wrap(. ~ year, ncol = 1)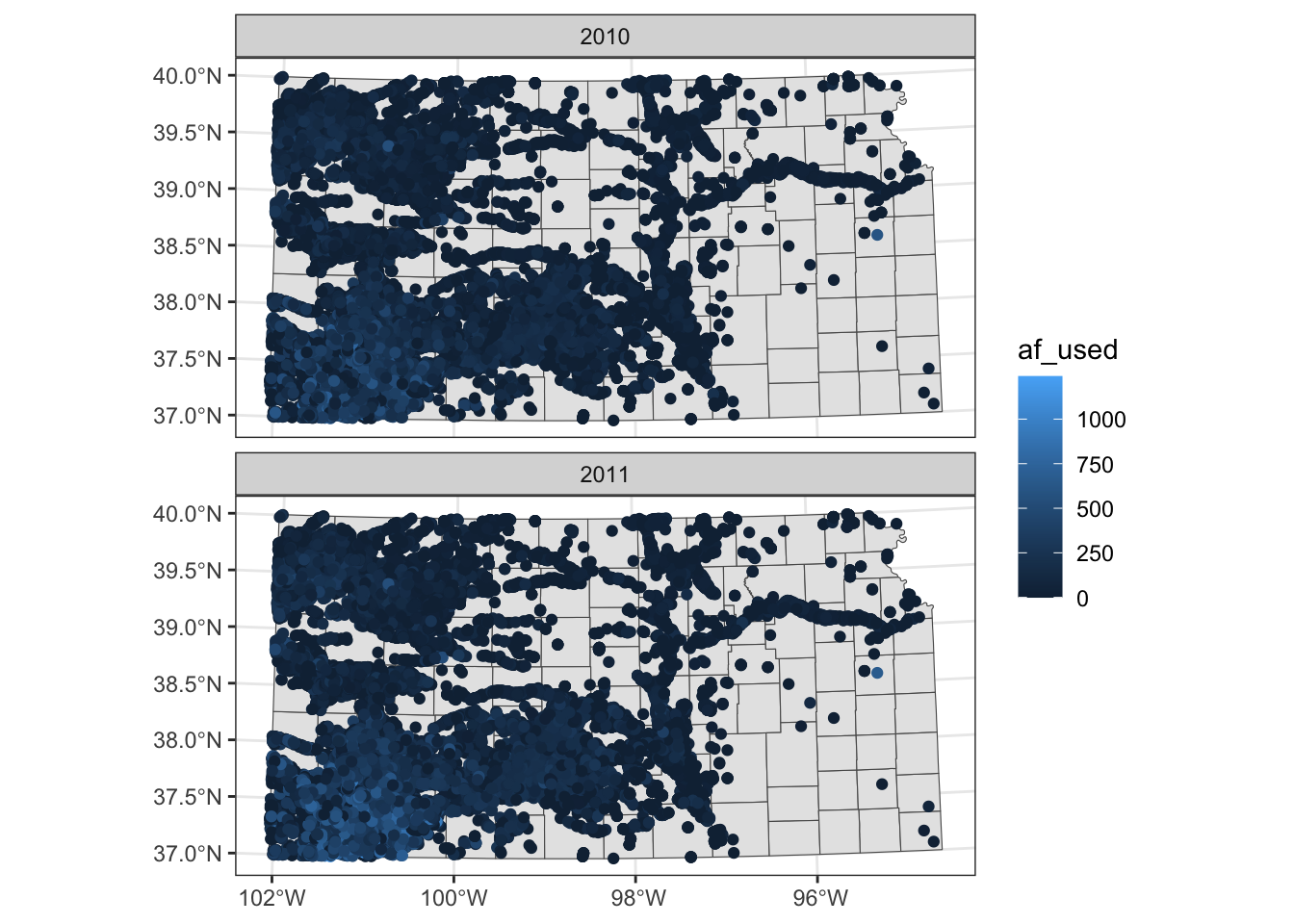
Two-way faceting is possible by supplying a variable name (or expression) in place of . in facet_wrap(. ~ year). The code below uses an expression (af_used > 200) in place of .. This divides the dataset by whether water use is greater than 200 or not and by year.
ggplot() +
#--- KS county boundary ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- wells ---#
geom_sf(data = gw_KS_sf, aes(color = af_used)) +
#--- facet by year (side by side) ---#
facet_wrap((af_used > 200) ~ year)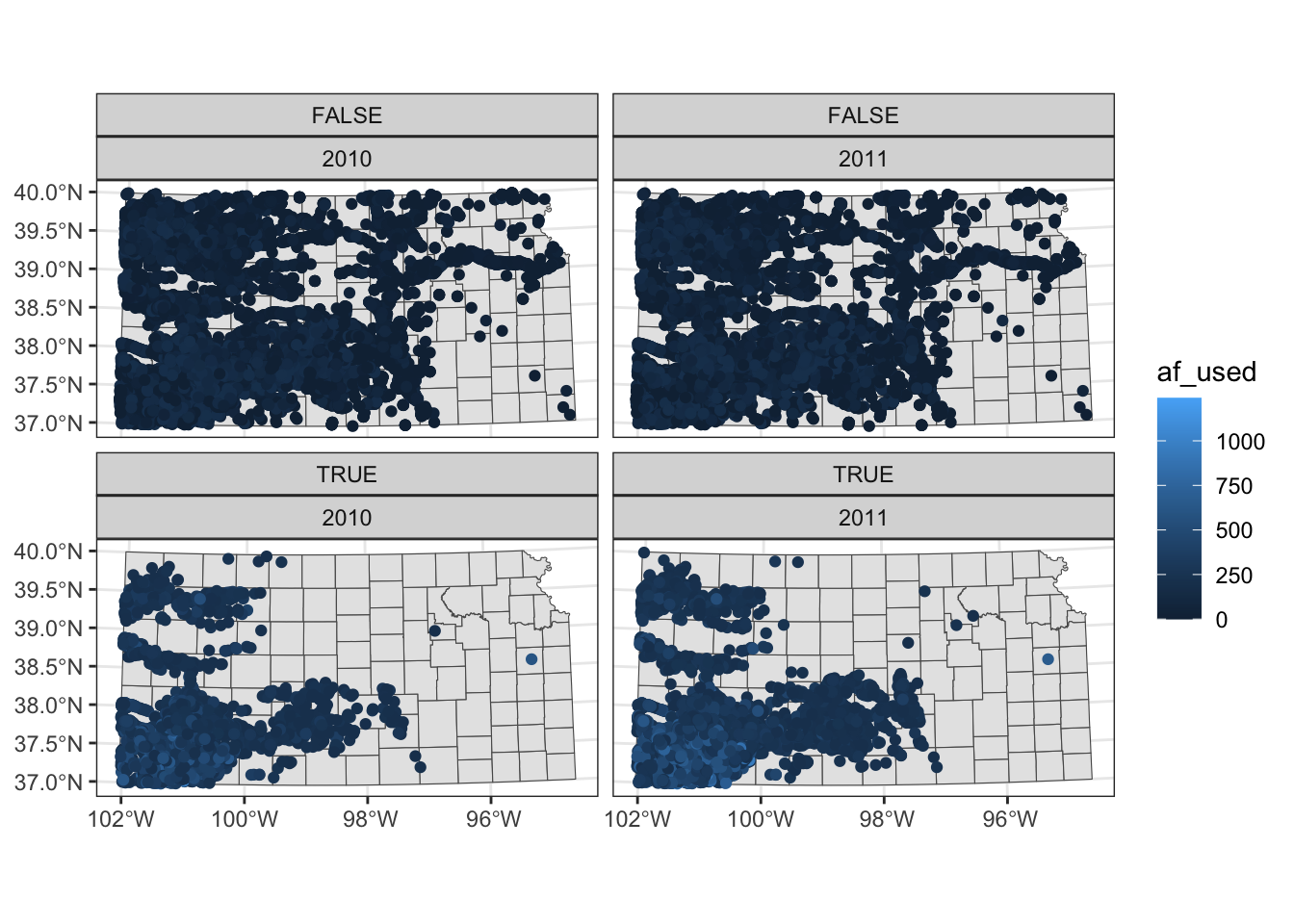
The values of the expression (TRUE or FALSE) appear in the gray strips, which is not informative. We will discuss in detail how to control texts in the strips section 8.5.
If you feel like the panels are too close to each other, you could provide more space between them using panel.spacing (both vertically and horizontally), panel.spacing.x (horizontally), and panel.spacing.y (vertically) options in theme(). Suppose you would like to place more space between the upper and lower panels, then you use panel.spacing.y like this:
ggplot() +
#--- KS county boundary ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- wells ---#
geom_sf(data = gw_KS_sf, aes(color = af_used)) +
#--- facet by year (side by side) ---#
facet_wrap((af_used > 200) ~ year) +
#--- add more space between panels ---#
theme(panel.spacing.y = unit(2, "lines"))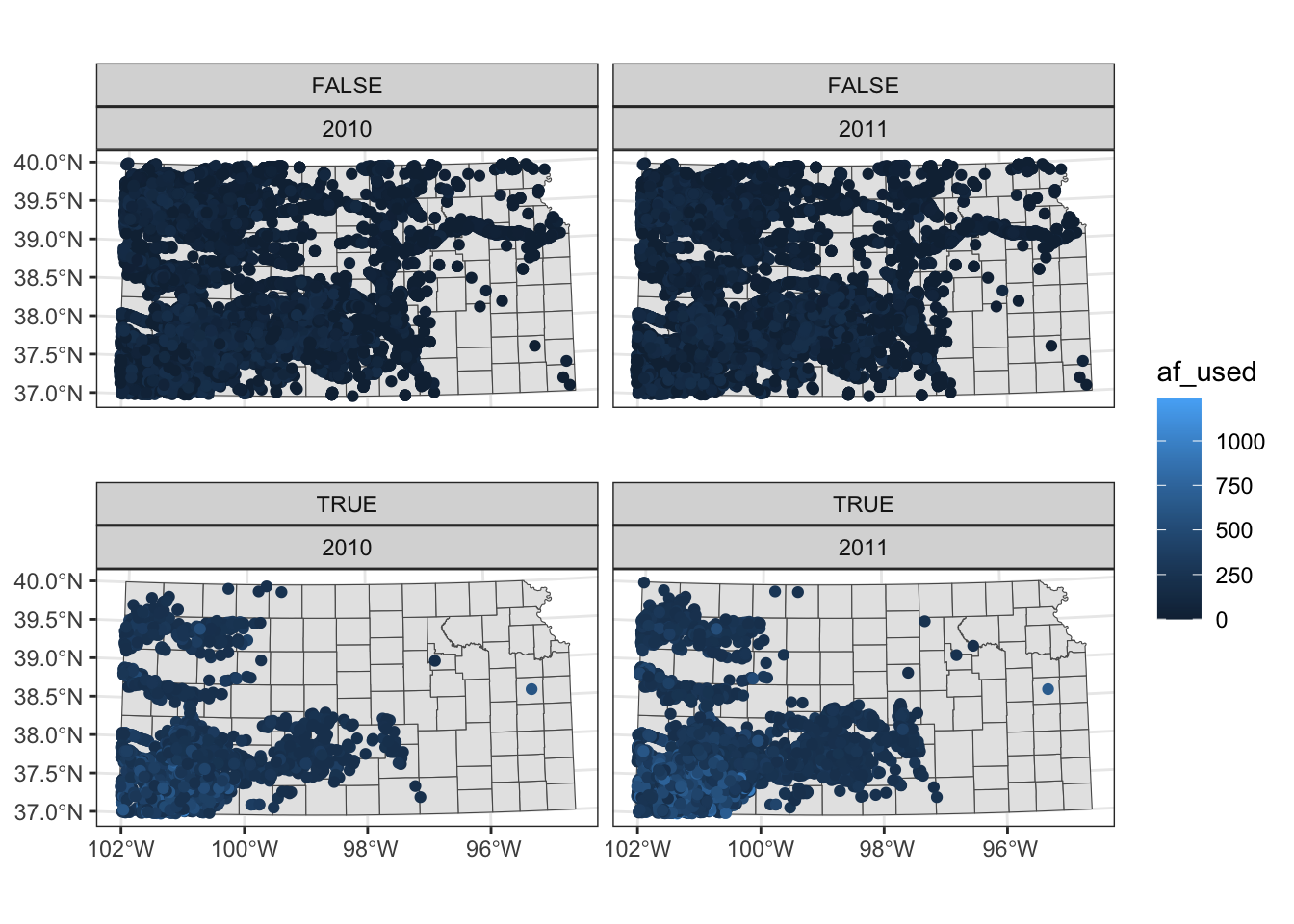
8.1.7 Adding texts (labels) on a map
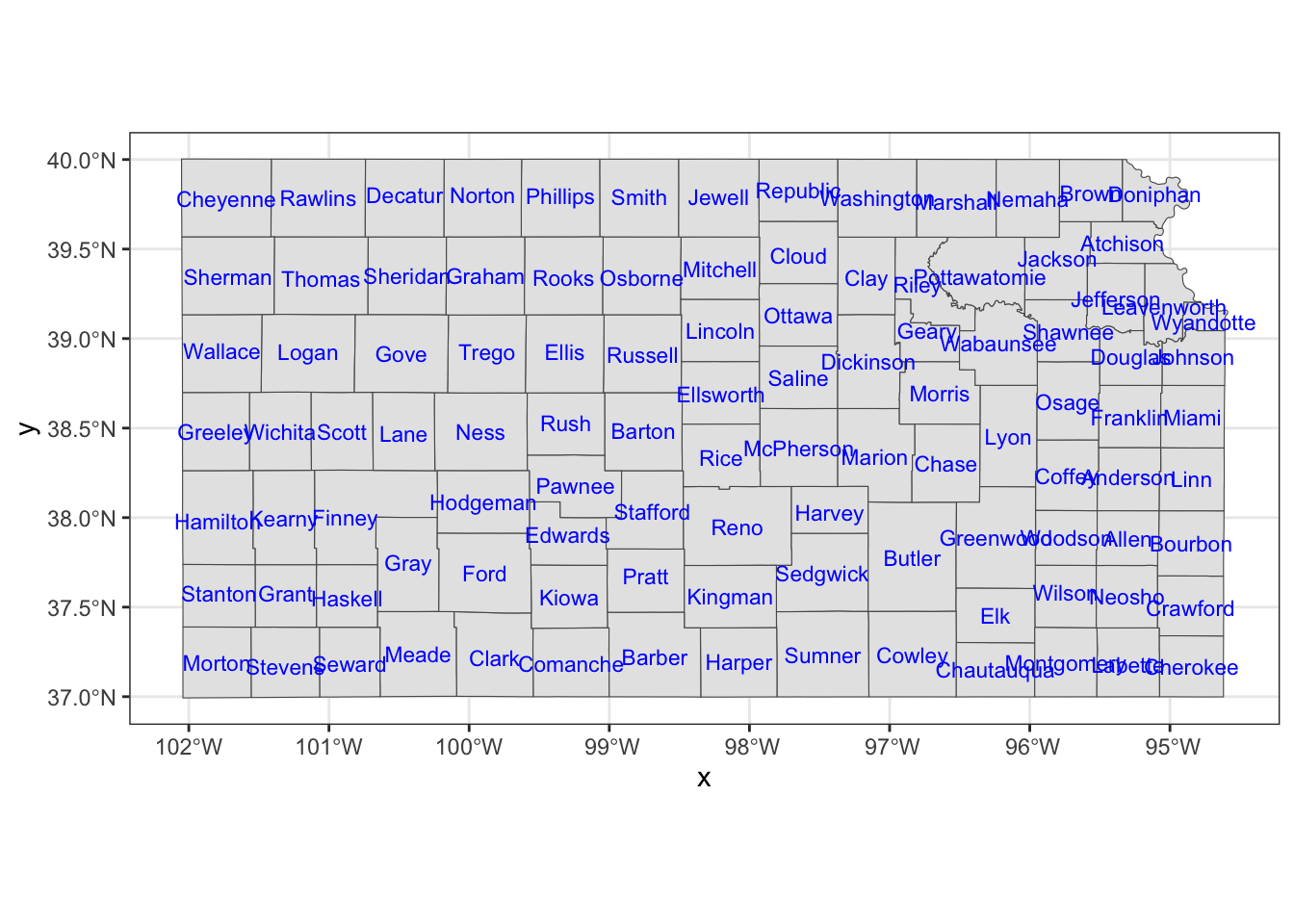
You can add labels to a map using geom_sf_text() or geom_sf_label() and providing aes(label = x) inside it where x is the variable that contains labels to print on a map.
ggplot() +
#--- KS county boundary ---#
geom_sf(data = KS_county) +
geom_sf_text(
data = KS_county,
aes(label = NAME),
size = 3,
color = "blue"
)
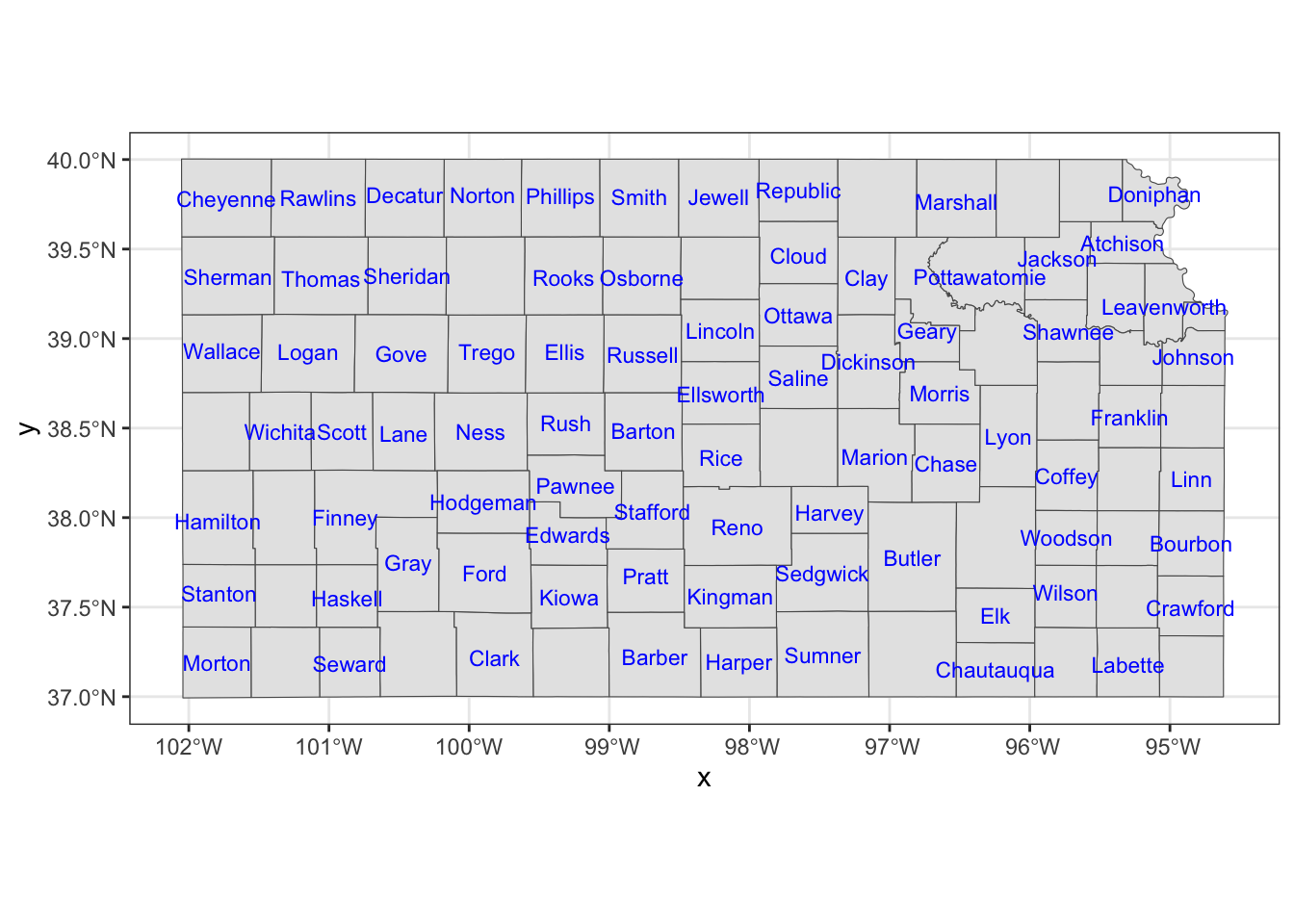
If you would like to have overlapping labels not printed, you can add check_overlap = TRUE.
ggplot() +
#--- KS county boundary ---#
geom_sf(data = KS_county) +
geom_sf_text(
data = KS_county,
aes(label = NAME),
check_overlap = TRUE,
size = 3,
color = "blue"
)
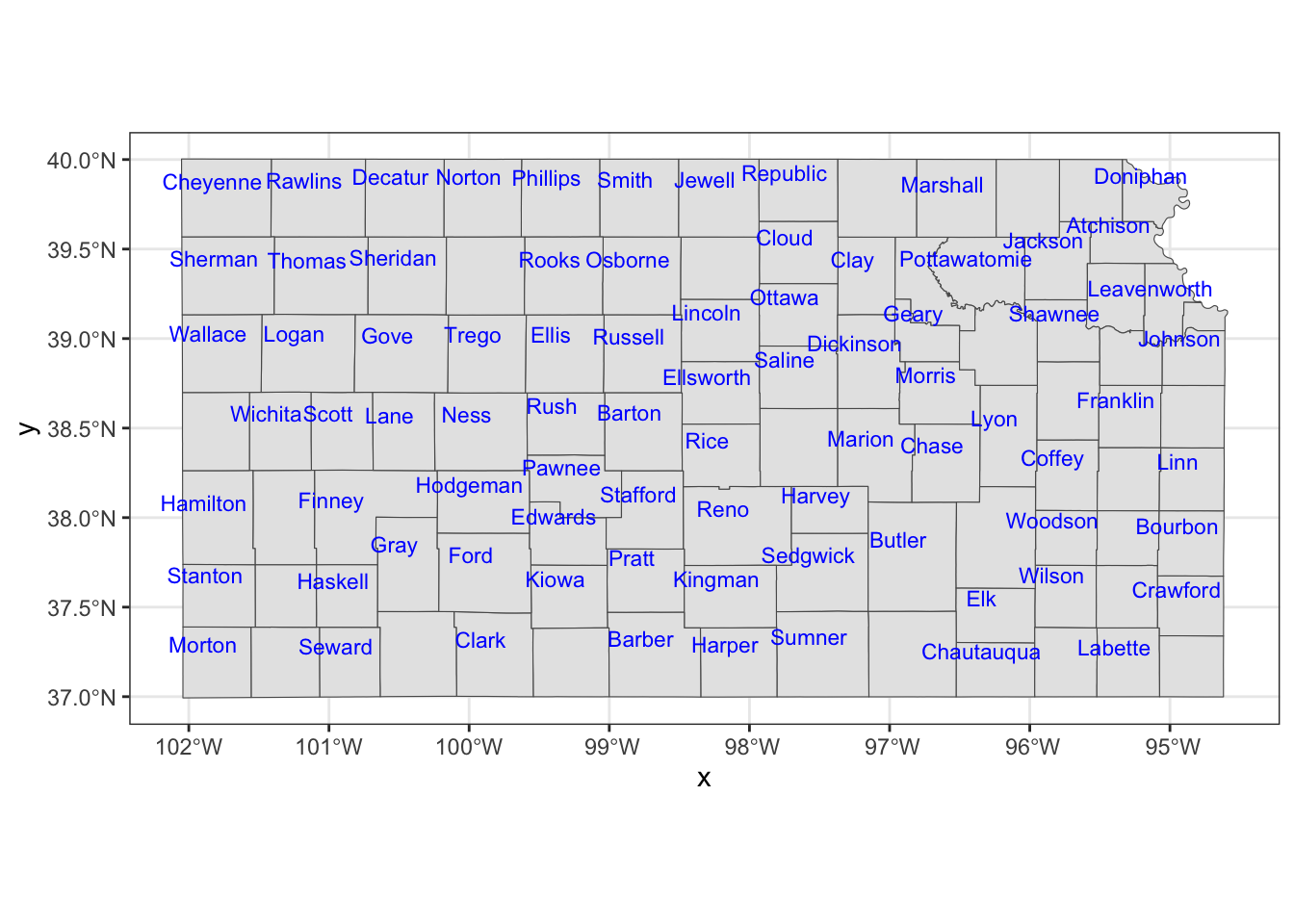
The nudge_x and nudge_y options let you shift the labels.
ggplot() +
#--- KS county boundary ---#
geom_sf(data = KS_county) +
geom_sf_text(
data = KS_county,
aes(label = NAME),
check_overlap = TRUE,
size = 3,
color = "blue",
nudge_x = -0.1,
nudge_y = 0.1
)
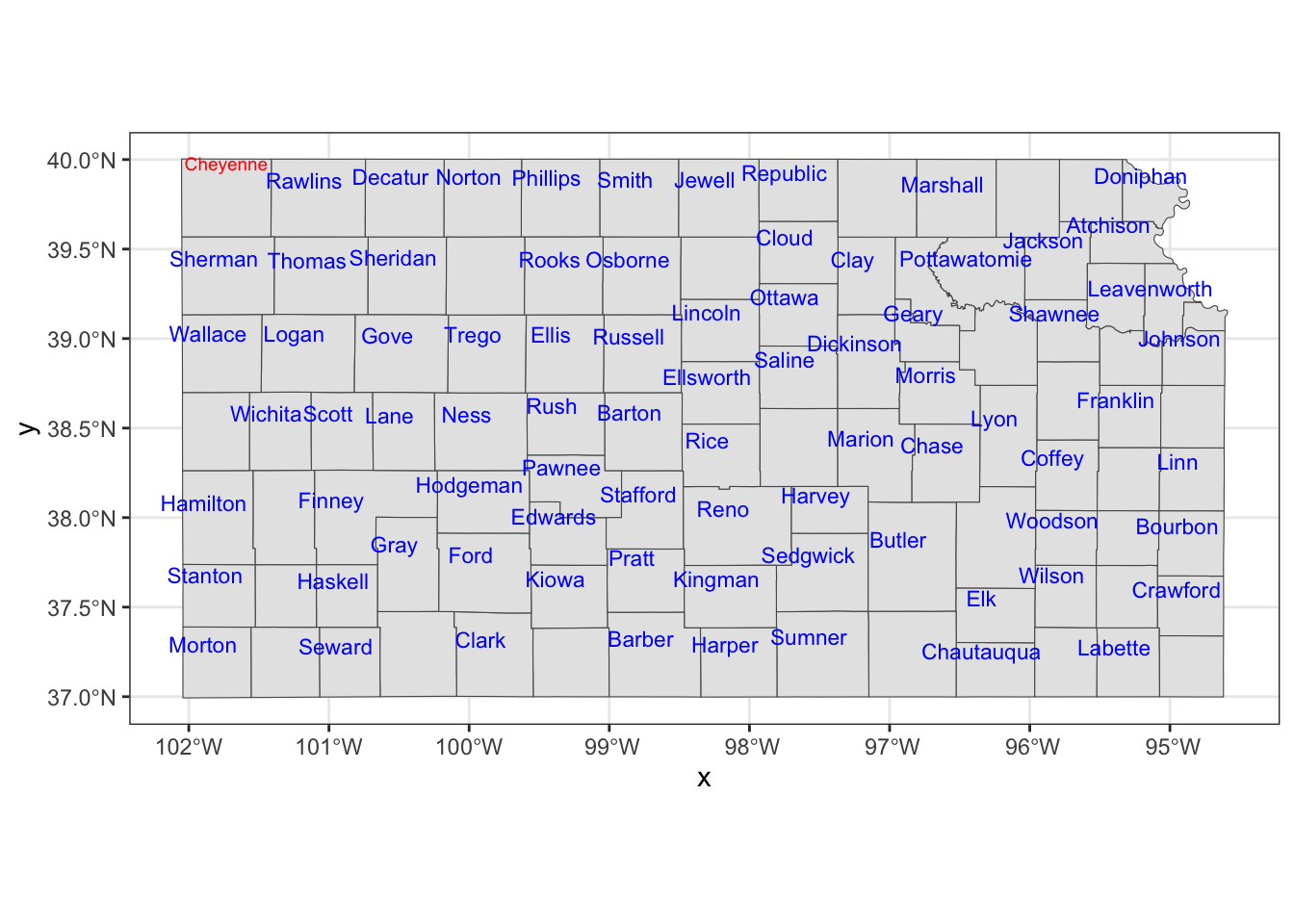
If you would like a fine control on a few objects, you can always work on them separately.
Cheyenne <- filter(KS_county, NAME == "Cheyenne")
KS_less_Cheyenne <- filter(KS_county, NAME != "Cheyenne")
ggplot() +
#--- KS county boundary ---#
geom_sf(data = KS_county) +
geom_sf_text(
data = KS_less_Cheyenne,
aes(label = NAME),
check_overlap = TRUE,
size = 3,
color = "blue",
nudge_x = -0.1,
nudge_y = 0.1
) +
geom_sf_text(
data = Cheyenne,
aes(label = NAME),
size = 2.5,
color = "red",
nudge_y = 0.2
)
You could also use annotate() to place texts on a map, which can be useful if you would like to place arbitrary texts that are not part of sf object.
ggplot() +
#--- KS county boundary ---#
geom_sf(data = KS_county) +
geom_sf_text(
data = KS_less_Cheyenne,
aes(label = NAME),
check_overlap = TRUE,
size = 3,
color = "blue",
nudge_x = -0.1,
nudge_y = 0.1
) +
#--- use annotate to add texts on the map ---#
annotate(
geom = "text",
x = -102,
y = 39.8,
size = 3,
label = "Cheyennes",
color = "red"
)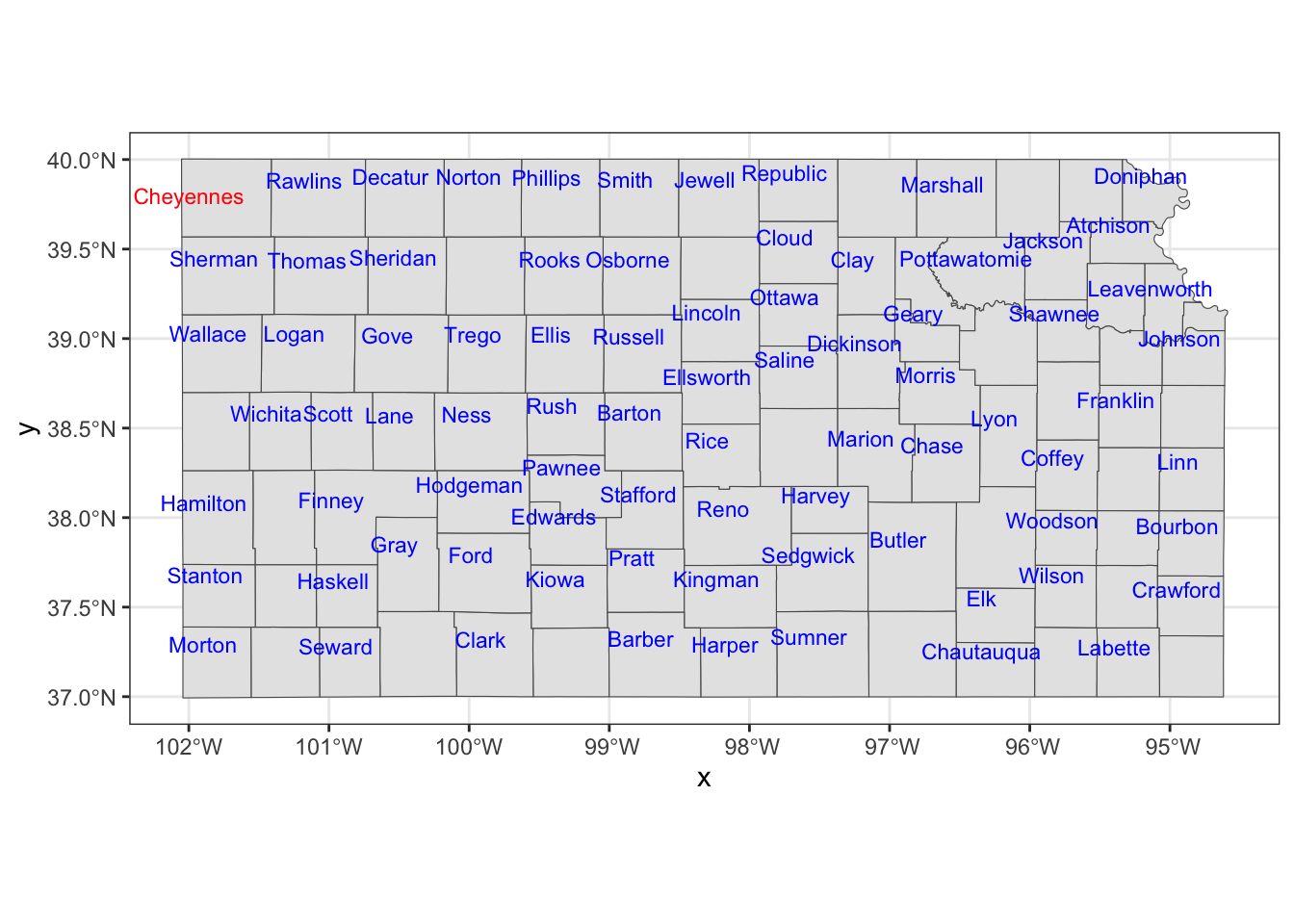
As you can see, you need to tell where the texts should be placed with x and y, provide the texts you want on the map to label.
8.2 Raster data visualization: geom_spatraster(), geom_raster(), and geom_stars()
This section shows how to use tidyterra::geom_spatraster(), ggplot2::geom_raster() and stars::geom_stars() to create maps from raster datasets of three object classes: SpatRaster from the terra pacakge, Raster\(^*\) objects from the raster package, and stars objects from the stars package, respectively.
Among these methods, geom_raster() does not accept SpatRaster, Raster$^*$, or stars objects as input. Instead, geom_raster() requires a data.frame containing coordinates to generate maps, making it a two-step process.
- convert raster dataset into a
data.framewith coordinates - use
geom_raster()to make a map
On the other hand, geom_spatraster() and geom_stars() accepts a SpatRaster (from the terra package) and stars object (from the stars package), respectivley, and no data transformation is necessary for either of them. For this reason, the use of geom_spatraster() or geom_stars is recommended over geom_raster().
8.2.1 Data Preparation
We use the following objects that have the same information but come in different object classes for illustration in this section.
Raster as stars
tmax_Jan_09 <- readRDS("Data/tmax_Jan_09_stars.rds")Raster as RasterStack
tmax_Jan_09_rs <- stack("Data/tmax_Jan_09.tif")Raster as SpatRaster
8.2.2 Visualize with tidyterra::geom_spatraster()
Creating a map from a SpatRaster is just as simple as creating a map from an sf object. You can simply use geom_spatraster() where the data argument is set to the SpatRaster object inside geom_spatraster(). To facet by layer, you can apply facet_wrap(~lyr) like below. Since it is under the ggplot2 framework, it is easy to tweak the figure as long as you are familiar with ggplot2.
ggplot() +
geom_spatraster(data = tmax_Jan_09_sr) +
facet_wrap(~ lyr, ncol = 2) +
scale_fill_whitebox_c(
palette = "muted",
labels = scales::label_number(suffix = "º"),
n.breaks = 20,
guide = guide_legend(reverse = TRUE)
) +
labs(fill = "")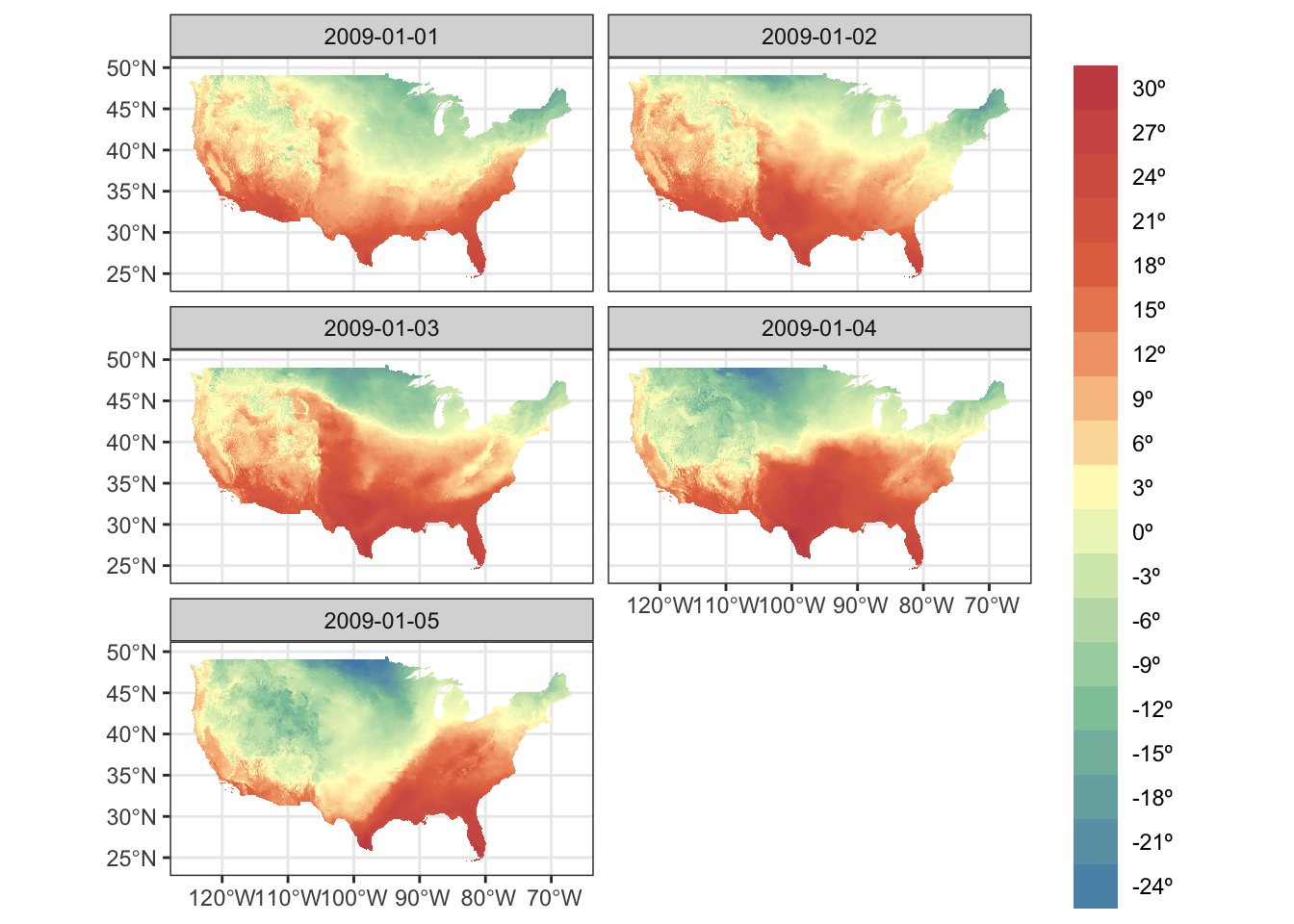
8.2.3 Visualize stars with geom_stars()
We saw above that geom_raster() requires converting a stars object to a data.frame first before creating a map. geom_stars() from the stars package lets you use a stars object directly to easily create a map under the ggplot2 framework. geom_stars() works just like geom_sf(). All you need to do is supply a stars object to geom_stars() as data.
ggplot() +
geom_stars(data = tmax_Jan_09) +
theme_void()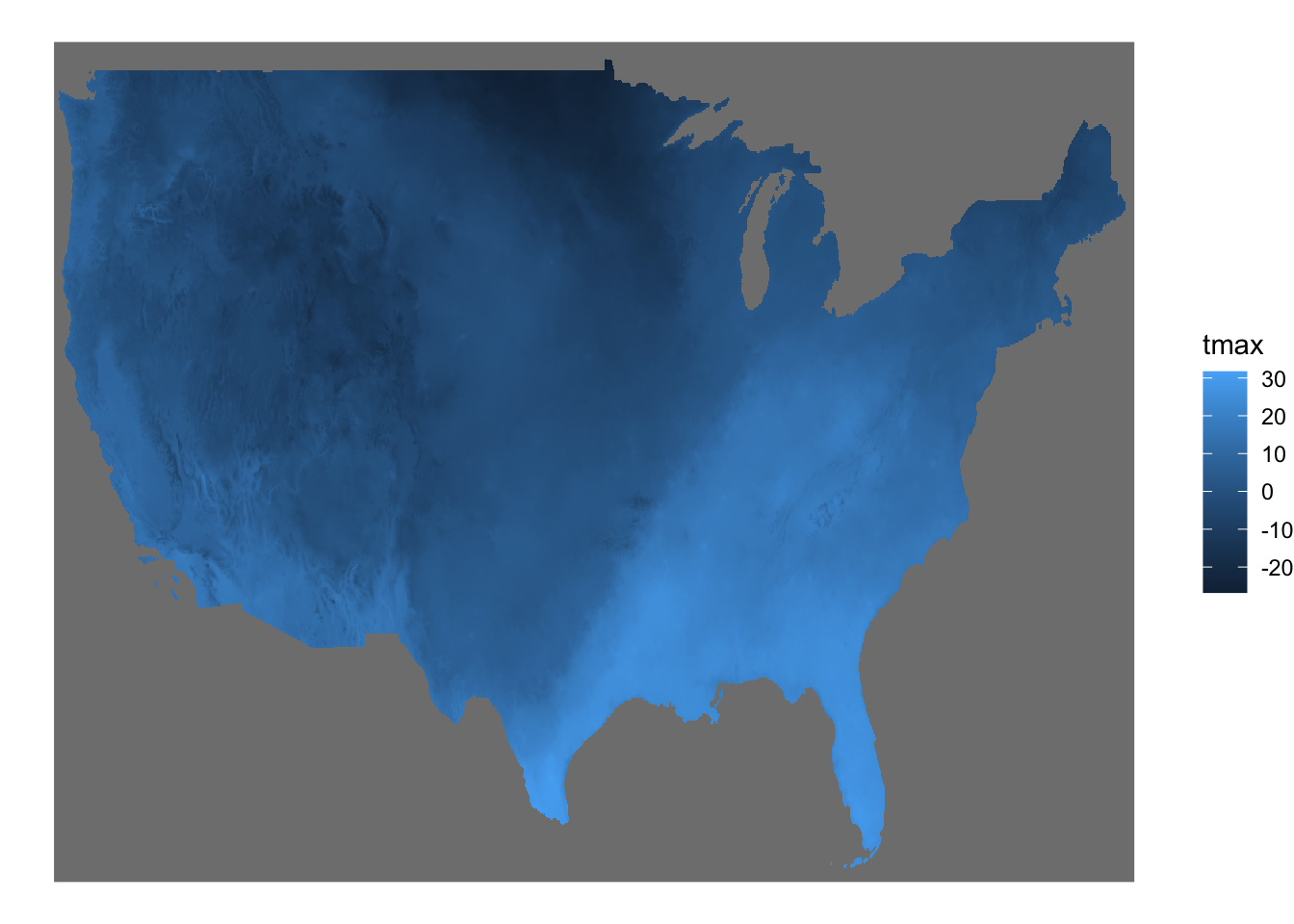
The fill color of the raster cells are automatically set to the attribute (here tmax) as if aes(fill = tmax). It is a good idea to add coord_equal() because of the same issue we saw with geom_raster().
ggplot() +
geom_stars(data = tmax_Jan_09) +
theme_void() +
coord_equal()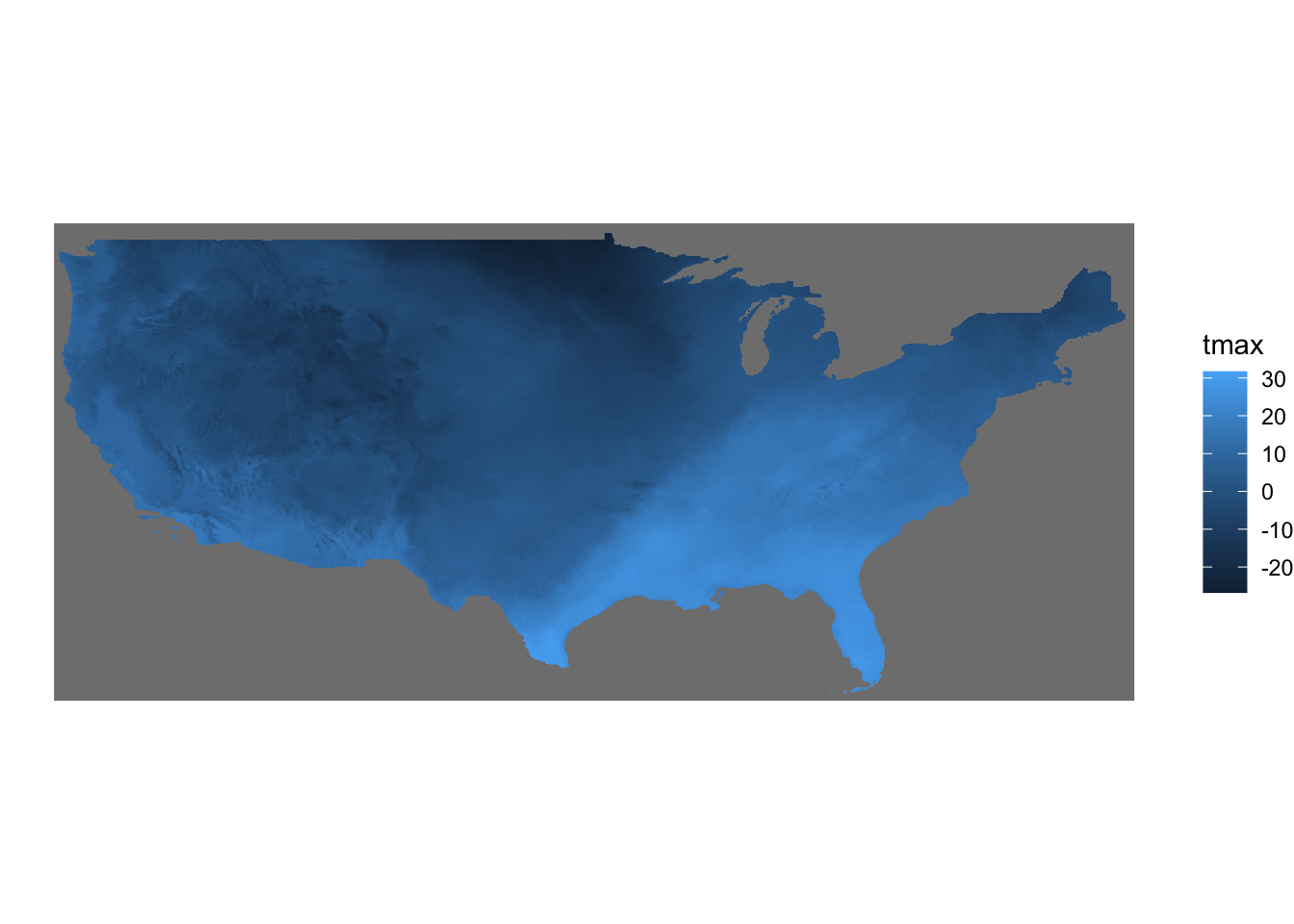
By default, geom_stars() plots only the first band. In order to present all the layers at the same time, you can add facet_wrap( ~ x) where x is the name of the third dimension of the stars object (date here).
ggplot() +
geom_stars(data = tmax_Jan_09) +
facet_wrap(~date) +
coord_equal() +
theme_void()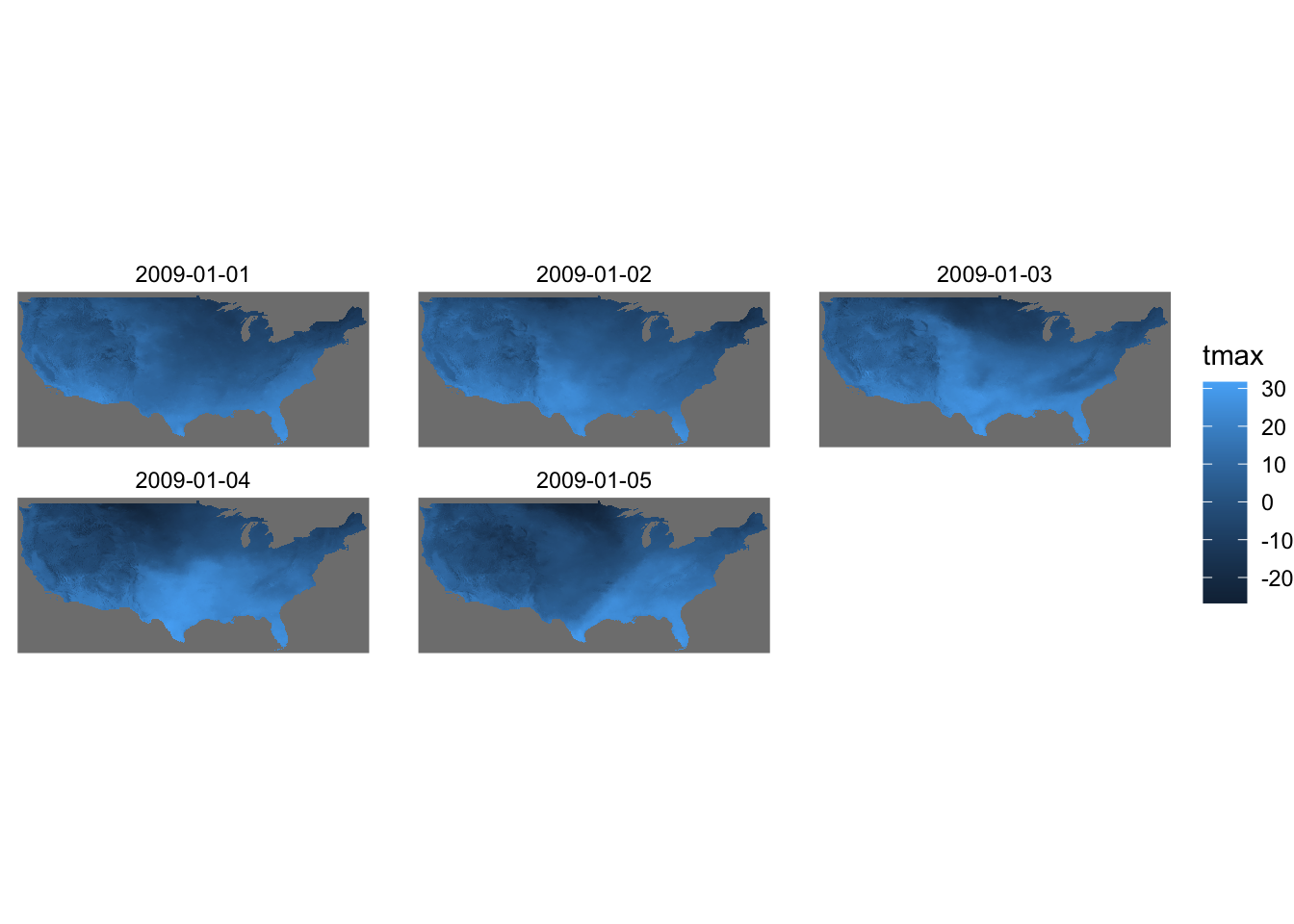
8.2.4 Visualize with geom_raster() (NOT recommended)
8.2.4.1 Visualize Raster\(^*\) with geom_raster()
In order to create maps from the information stored in Raster\(^*\) objects, you first convert them to a regular data.frame using as.data.frame() as follows:
#--- convert to data.frame ---#
tmax_Jan_09_df <-
as.data.frame(tmax_Jan_09_rs, xy = TRUE) %>%
#--- remove cells with NA for any of the layers ---#
na.omit() %>%
#--- change the variable names ---#
setnames(
paste0("layer.", 1:5),
seq(ymd("2009-01-01"), ymd("2009-01-05"), by = "days") %>%
as.character()
)
#--- take a look ---#
head(tmax_Jan_09_df) x y 2009-01-01 2009-01-02 2009-01-03 2009-01-04 2009-01-05
17578 -95.12500 49.41667 -12.863 -11.455 -17.516 -12.755 -26.446
18982 -95.16667 49.37500 -12.698 -11.441 -17.436 -12.672 -25.889
18983 -95.12500 49.37500 -12.921 -11.611 -17.589 -12.830 -26.544
18984 -95.08333 49.37500 -13.074 -11.827 -17.684 -12.913 -26.535
18985 -95.04167 49.37500 -13.591 -12.220 -17.920 -12.684 -25.780
18986 -95.00000 49.37500 -13.688 -12.177 -17.911 -12.548 -25.424The xy = TRUE option adds the coordinates of the centroid of the raster cells, and na.omit() removes cells that are outside of the Kansas border and have NA values. Notice that each band comprises a column in the data.frame.
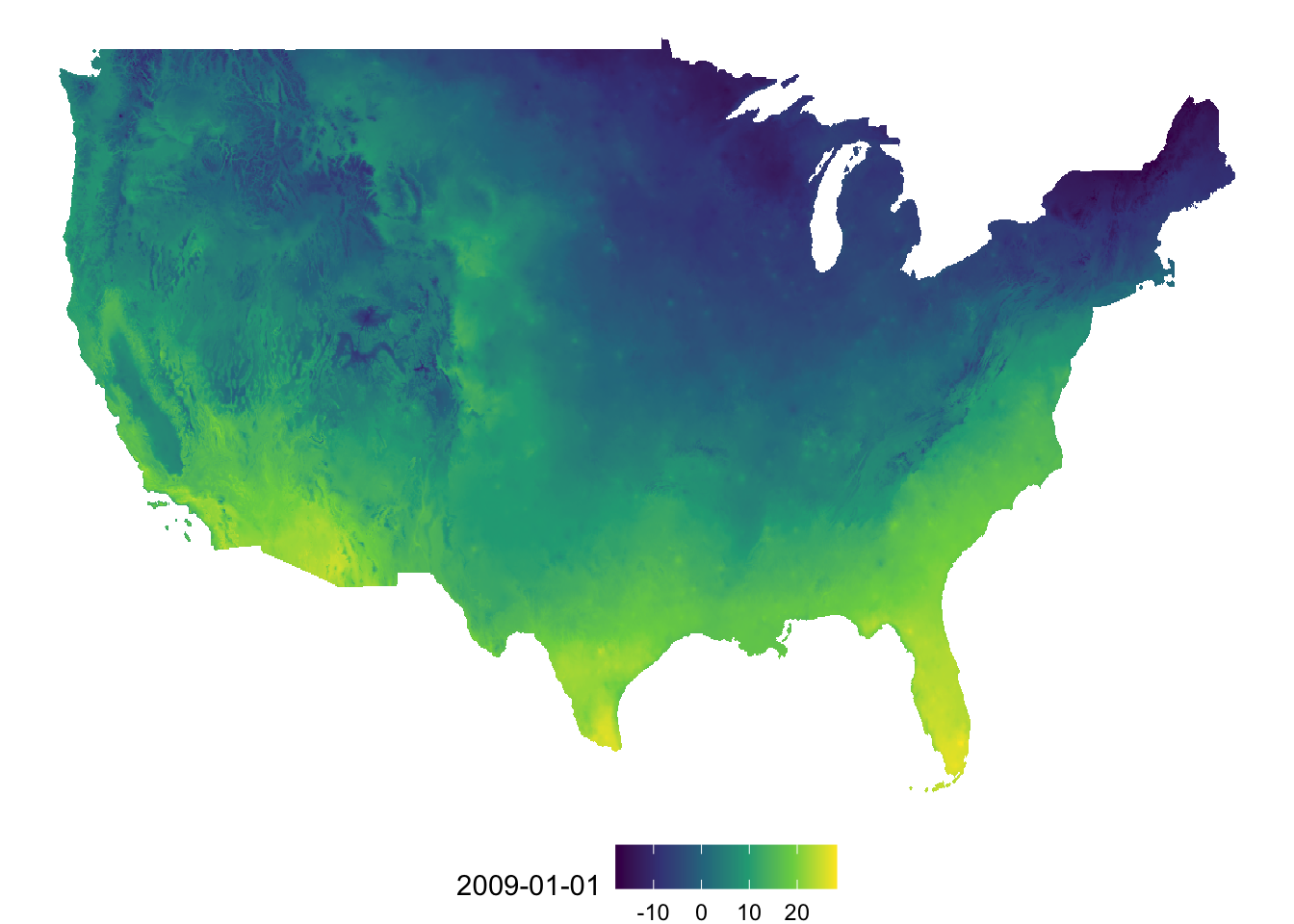
Once the conversion is done, you can use geom_raster() to create a map. Unlike geom_sf(), you need to supply the variables names for the geographical coordinates (here x for longitude and y for latitude). Further, you also need to specify which variable to use for fill color differentiation.
(
g_tmax_map <- ggplot(data = tmax_Jan_09_df) +
geom_raster(aes(x = x, y = y, fill = `2009-01-01`)) +
scale_fill_viridis_c() +
theme_void() +
theme(
legend.position = "bottom"
)
)
You can add coord_equal() so that one degree in latitude and longitude are the same length on the map. Of course, one degree in longitude and one degree in latitude are not the same length for US. However, distortion is smaller compared to the default at least.
g_tmax_map + coord_equal()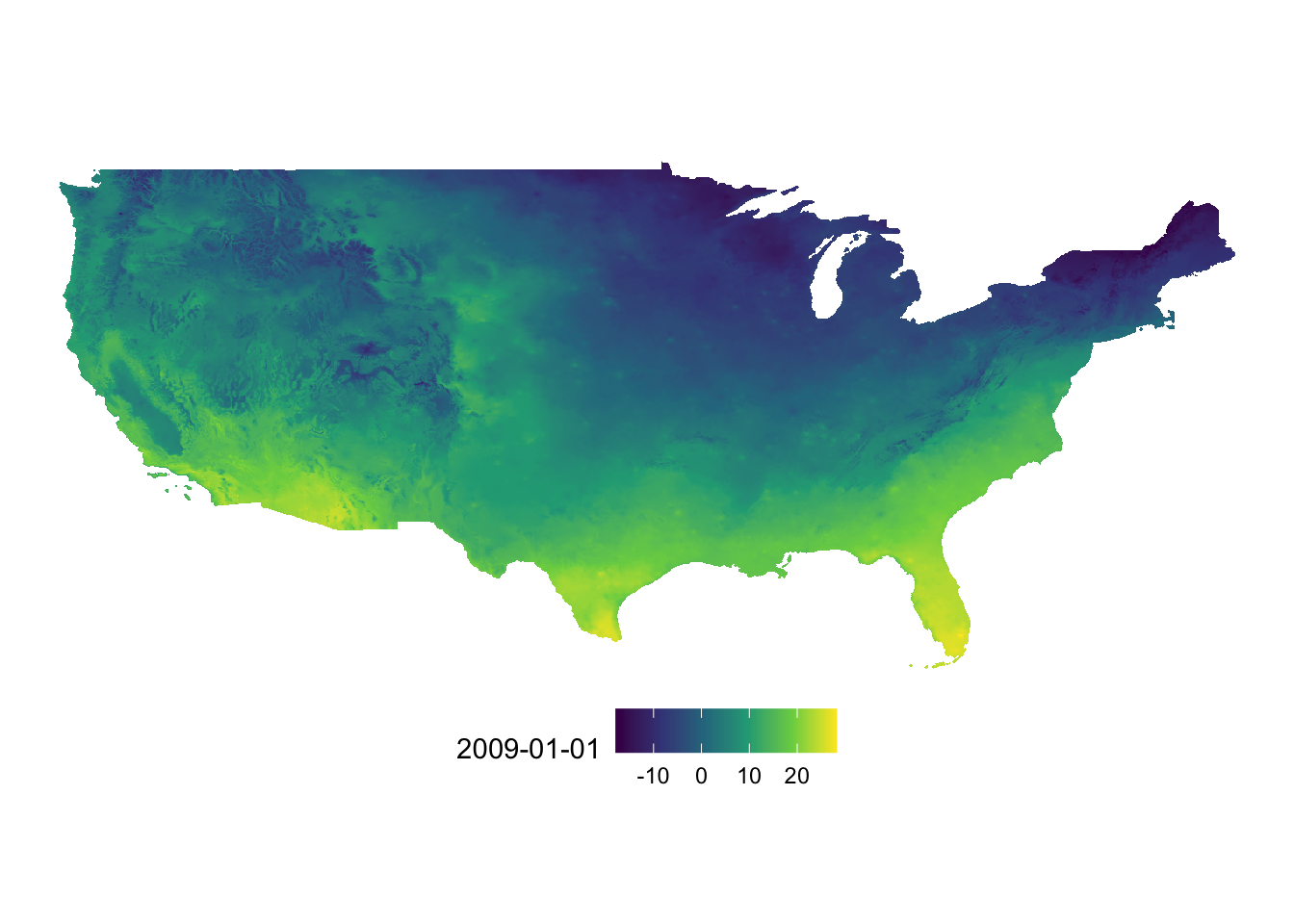
We only visualized tmax data for one day (layer.1) out of five days of tmax records. To present all of them at the same time we can facet using facet_wrap(). However, before we do that we need to have the data in a long format like this:
tmax_long_df <-
tmax_Jan_09_df %>%
pivot_longer(
c(-x, -y),
names_to = "date",
values_to = "tmax"
)
#--- take a look ---#
head(tmax_long_df)# A tibble: 6 × 4
x y date tmax
<dbl> <dbl> <chr> <dbl>
1 -95.1 49.4 2009-01-01 -12.9
2 -95.1 49.4 2009-01-02 -11.5
3 -95.1 49.4 2009-01-03 -17.5
4 -95.1 49.4 2009-01-04 -12.8
5 -95.1 49.4 2009-01-05 -26.4
6 -95.2 49.4 2009-01-01 -12.7Let’s now use facet_wrap() to create a series of tmax maps.
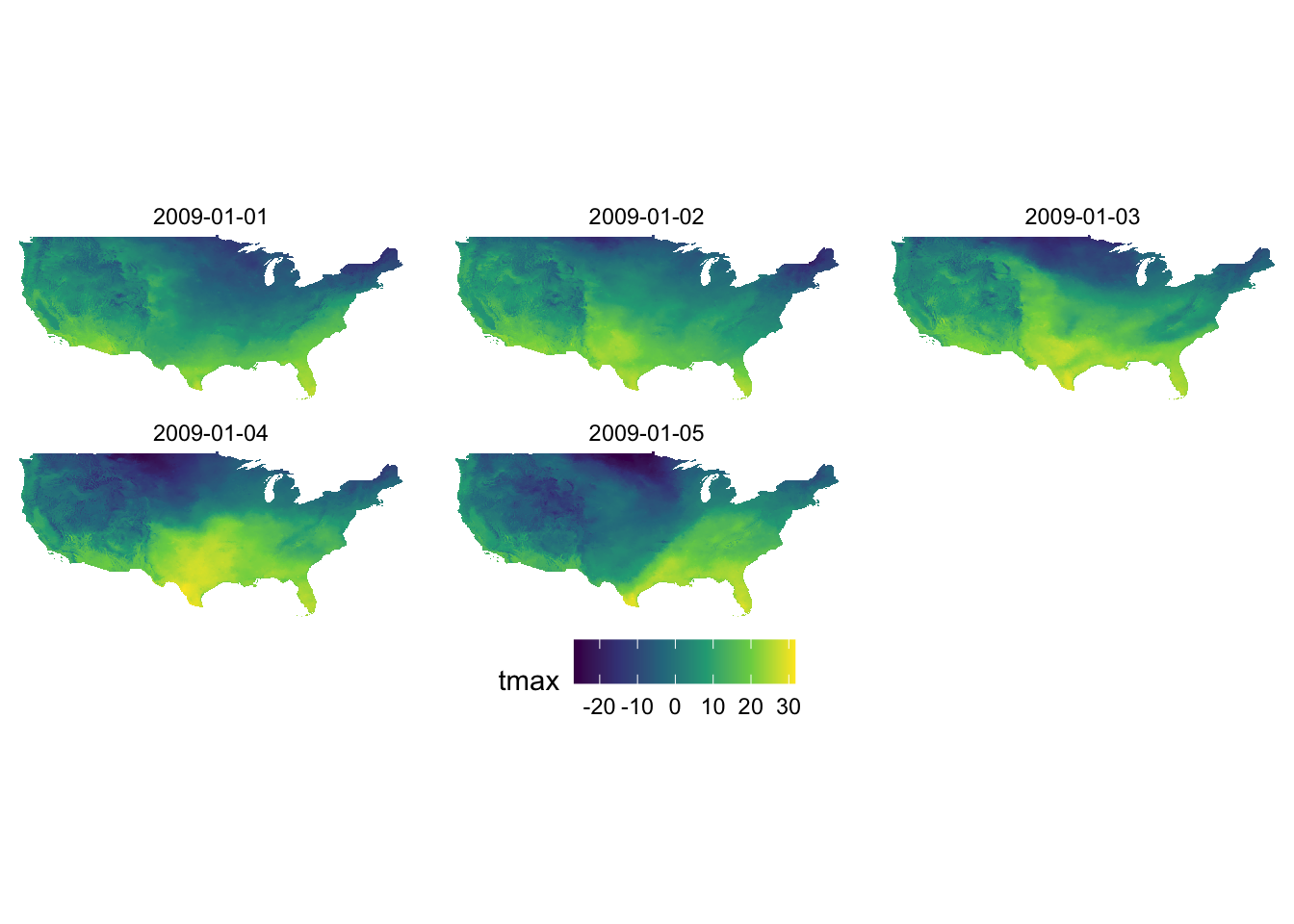
ggplot() +
geom_raster(data = tmax_long_df, aes(x = x, y = y, fill = tmax)) +
facet_wrap(date ~ .) +
coord_equal() +
scale_fill_viridis_c() +
theme_void() +
theme(
legend.position = "bottom"
)
8.2.4.2 Visualize stars with geom_raster()
Similarly with Raster\(^*\) objects, we first need to convert a stars to a regular data.frame using as.data.frame() as follows:
#--- converting to a data.frame ---#
tmax_long_df <-
as.data.frame(tmax_Jan_09, xy = TRUE) %>%
na.omit()
#--- take a look ---#
head(tmax_Jan_09_df) x y 2009-01-01 2009-01-02 2009-01-03 2009-01-04 2009-01-05
17578 -95.12500 49.41667 -12.863 -11.455 -17.516 -12.755 -26.446
18982 -95.16667 49.37500 -12.698 -11.441 -17.436 -12.672 -25.889
18983 -95.12500 49.37500 -12.921 -11.611 -17.589 -12.830 -26.544
18984 -95.08333 49.37500 -13.074 -11.827 -17.684 -12.913 -26.535
18985 -95.04167 49.37500 -13.591 -12.220 -17.920 -12.684 -25.780
18986 -95.00000 49.37500 -13.688 -12.177 -17.911 -12.548 -25.424One key difference from the conversion of Raster\(^*\) objects is that the data.frame is already in the long format, which means that you can immediately make faceted figures like this:
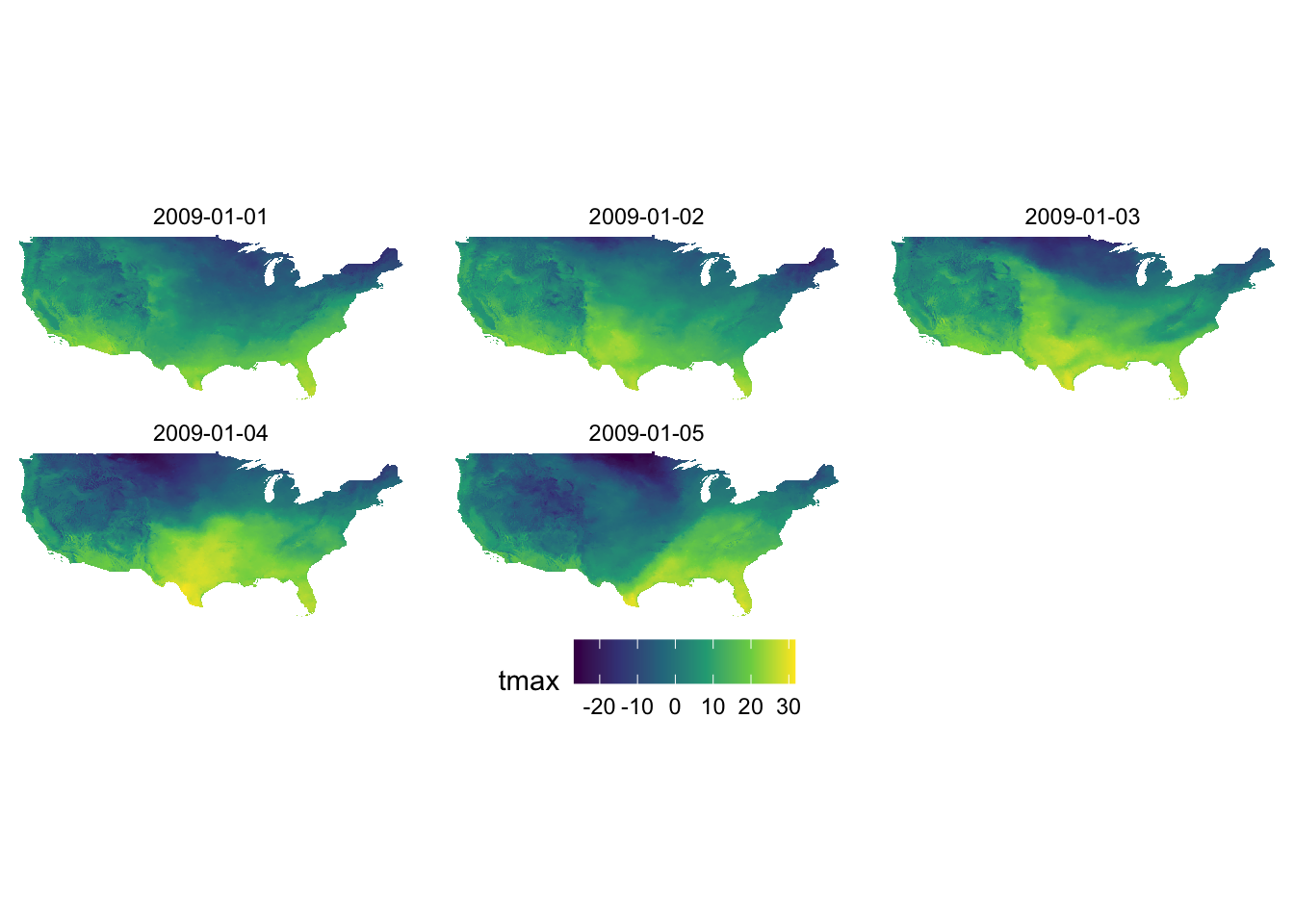
ggplot() +
geom_raster(data = tmax_long_df, aes(x = x, y = y, fill = tmax)) +
facet_wrap(date ~ .) +
coord_equal() +
scale_fill_viridis_c() +
theme_void() +
theme(
legend.position = "bottom"
)
8.2.5 adding geom_sf() layers
You can easily add geom_sf() layers to a map created with any of geom_spatraster(), geom_stars(), and geom_raster(). Let’s crop the tmax data to Kansas and create a map of tmax values displayed on top of the Kansas county borders.
#--- crop to KS ---#
KS_tmax_Jan_09_stars <-
tmax_Jan_09 %>%
st_crop(., KS_county)
#--- convert to a df ---#
KS_tmax_Jan_09_df <-
as.data.frame(KS_tmax_Jan_09_stars, xy = TRUE) %>%
na.omit()
8.2.5.1 adding geom_sf() to a map with geom_raster()
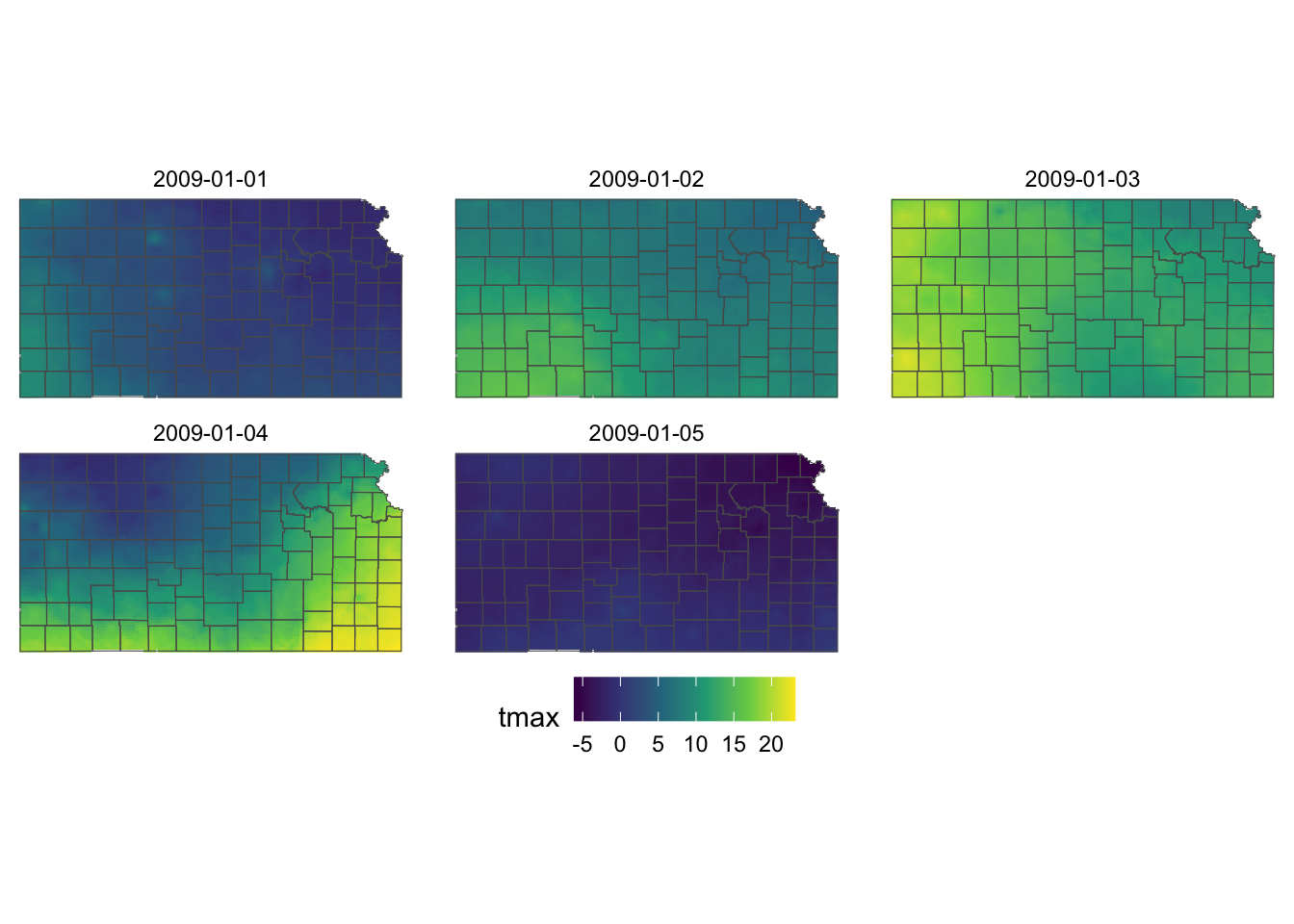
ggplot() +
geom_raster(data = KS_tmax_Jan_09_df, aes(x = x, y = y, fill = tmax)) +
geom_sf(data = KS_county, fill = NA) +
facet_wrap(date ~ .) +
scale_fill_viridis_c() +
theme_void() +
theme(
legend.position = "bottom"
)
Notice that coord_equal() is not necessary in the above code. Indeed, if you try to add coord_equal(), you will have an error.
ggplot() +
geom_raster(data = KS_tmax_Jan_09_df, aes(x = x, y = y, fill = tmax)) +
geom_sf(data = KS_county, fill = NA) +
facet_wrap(date ~ .) +
coord_equal() +
scale_fill_viridis_c() +
theme_void() +
theme(
legend.position = "bottom"
)Error in `geom_sf()`:
! Problem while converting geom to grob.
ℹ Error occurred in the 2nd layer.
Caused by error in `draw_panel()`:
! `geom_sf()` can only be used with `coord_sf()`.Further, the original stars or Raster\(^*\) must have the same CRS as the sf objects. The following code transforms the CRS of KS_county to 32614 and try to plot them together.
ggplot() +
geom_raster(data = KS_tmax_Jan_09_df, aes(x = x, y = y, fill = tmax)) +
geom_sf(data = st_transform(KS_county, 32614), fill = NA) +
facet_wrap(date ~ .) +
scale_fill_viridis_c() +
theme_void() +
theme(
legend.position = "bottom"
)
When plotting multiple sf objects, the CRS for the map was set to the CRS of the sf objects used for the first geom_sf() layer, and the rest of the sf objects followed suit. That is not the case here.
8.2.5.2 adding geom_sf() to a map with geom_stars()
This is basically the same as the case with geom_stars()
ggplot() +
geom_stars(data = KS_tmax_Jan_09_stars) +
geom_sf(data = KS_county, fill = NA) +
facet_wrap(~date) +
theme_void()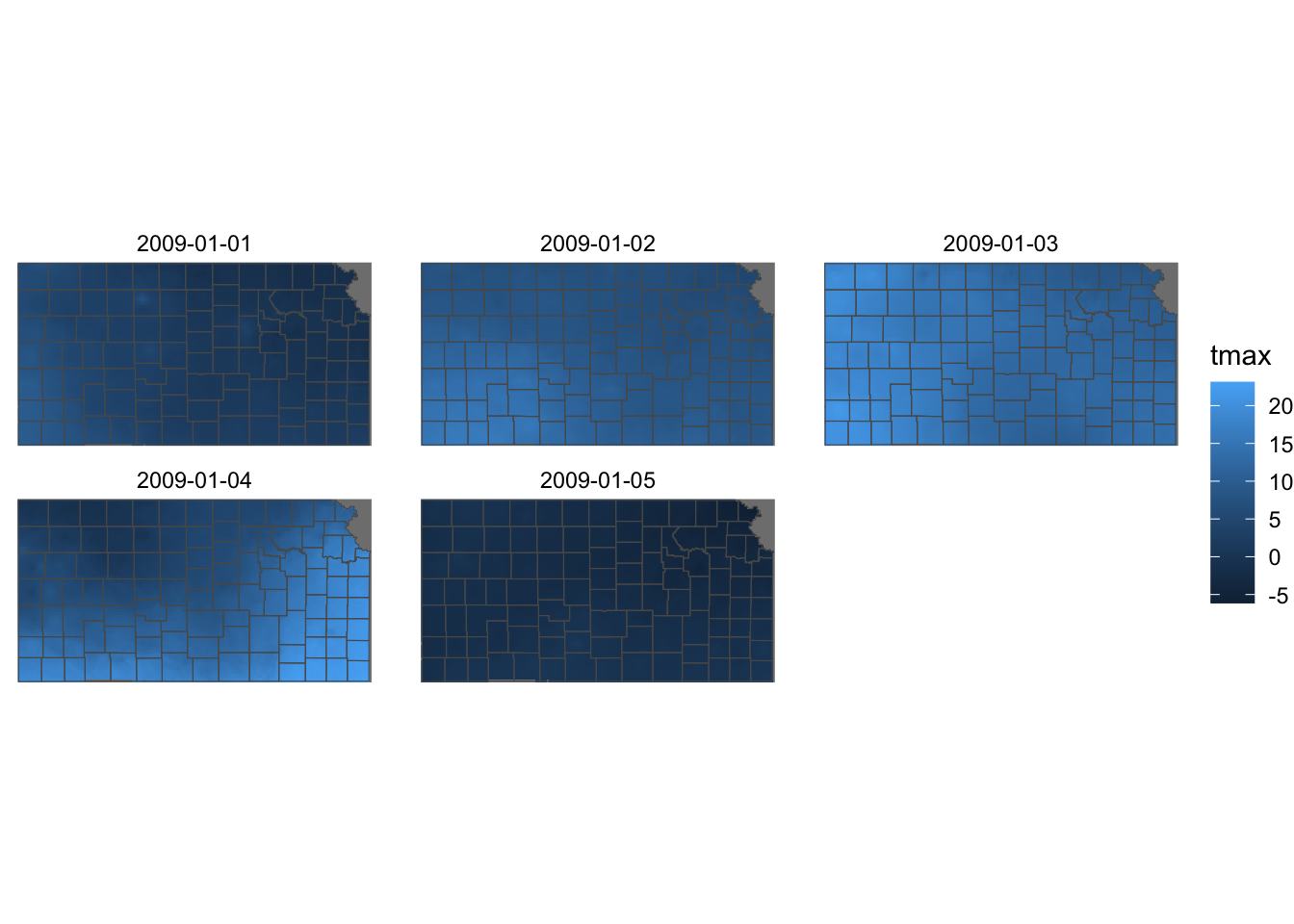
Just like the case with geom_raster(), coord_equal() is not necessary and the stars and sf objects must have the same CRS.
ggplot() +
geom_stars(data = KS_tmax_Jan_09_stars) +
geom_sf(data = st_transform(KS_county, 32614), fill = NA) +
facet_wrap(~date) +
theme_void()
8.3 Color scale
Color scale refers to the way variable values are mapped to colors on a figure. For example, in the example below, the color scale for fill maps the value of af_used to a gradient of colors: dark blue for low values to light blue for high values of af_used.
tmax_long_df <- readRDS("Data/tmax_Jan_09_stars.rds") %>%
as.data.frame(xy = TRUE) %>%
na.omit()
g_col_scale <- ggplot() +
geom_raster(data = tmax_long_df, aes(x = x, y = y, fill = tmax)) +
facet_wrap(date ~ .) +
coord_equal() +
theme_void() +
theme(
legend.position = "bottom"
)Often times, it is aesthetically desirable to change the default color scale of ggplot2. For example, if you would like to color-differentiate temperature values, you might want to start from blue for low values to red for high values.
You can control the color scale using scale_*() functions. Which scale_*() function to use depends on the type of aesthetics (fill or color) and whether the aesthetic variable is continuous or discrete. Providing a wrong kind of scale_*() function results in an error.
8.3.1 Viridis color maps
The ggplot2 packages offers scale_A_viridis_B() functions for viridis color map, where A is the type of aesthetics attribute (fill, color), and B is the type of variable. For example, scale_fill_viridis_c() can be used for fill aesthetics applied to a continuous variable.
There are five types of palettes available under the viridis color map and can be selected using the option = option. Here is a visualization of all the five palettes.
data("geyser", package = "MASS")
ggplot(geyser, aes(x = duration, y = waiting)) +
xlim(0.5, 6) +
ylim(40, 110) +
stat_density2d(aes(fill = ..level..), geom = "polygon") +
theme_bw() +
theme(panel.grid = element_blank()) -> gg
(gg + scale_fill_viridis_c(option = "A") + labs(x = "magma", y = NULL)) /
(gg + scale_fill_viridis_c(option = "B") + labs(x = "inferno", y = NULL)) /
(gg + scale_fill_viridis_c(option = "C") + labs(x = "plasma", y = NULL)) /
(gg + scale_fill_viridis_c(option = "D") + labs(x = "viridis", y = NULL)) /
(gg + scale_fill_viridis_c(option = "E") + labs(x = "cividis", y = NULL))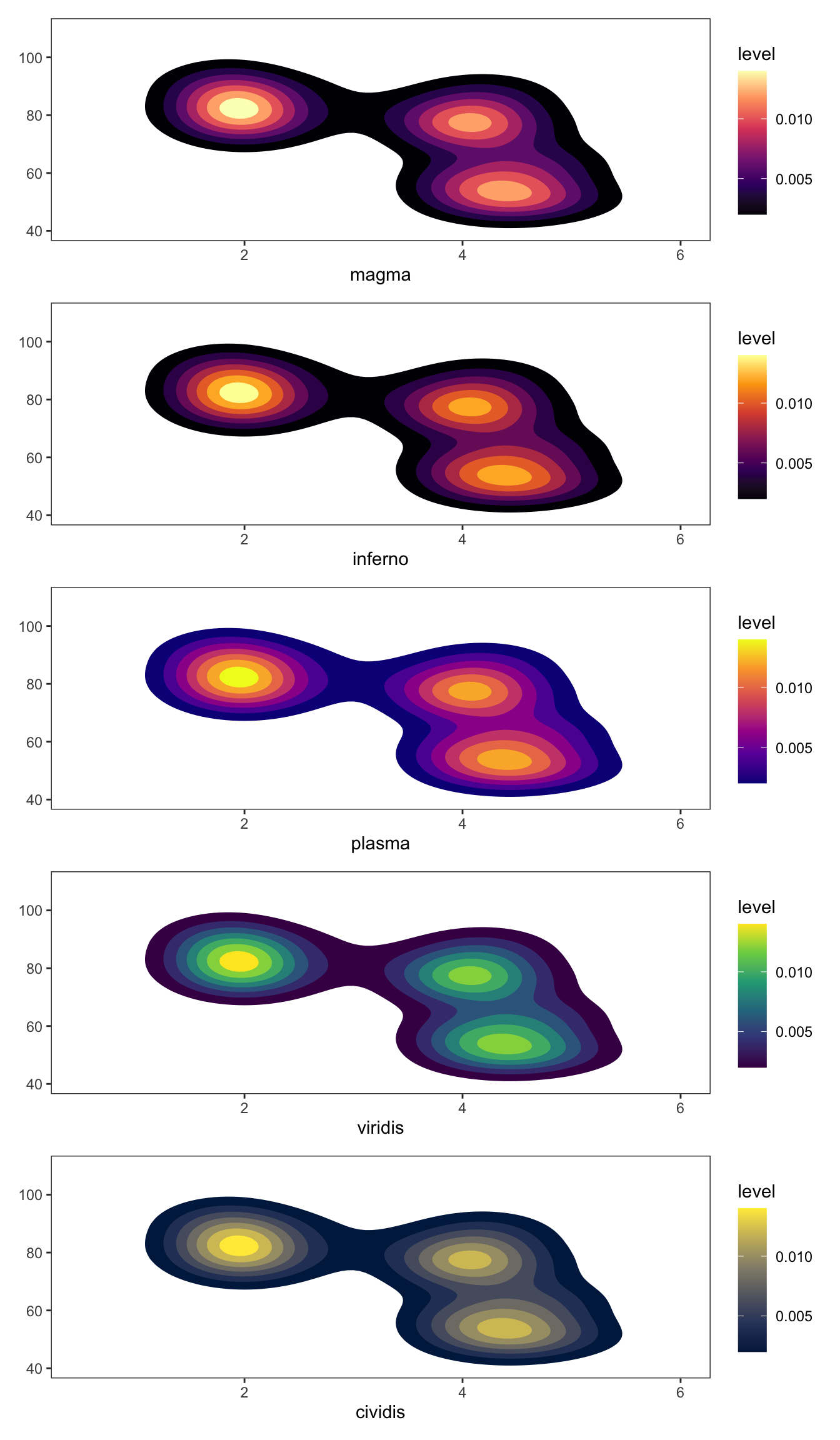
Let’s see what the PRISM tmax maps look like using Option A and D (default). Since the aesthetics type is fill and tmax is continuous, scale_fill_viridis_c() is the appropriate one here.
g_col_scale + scale_fill_viridis_c(option = "A")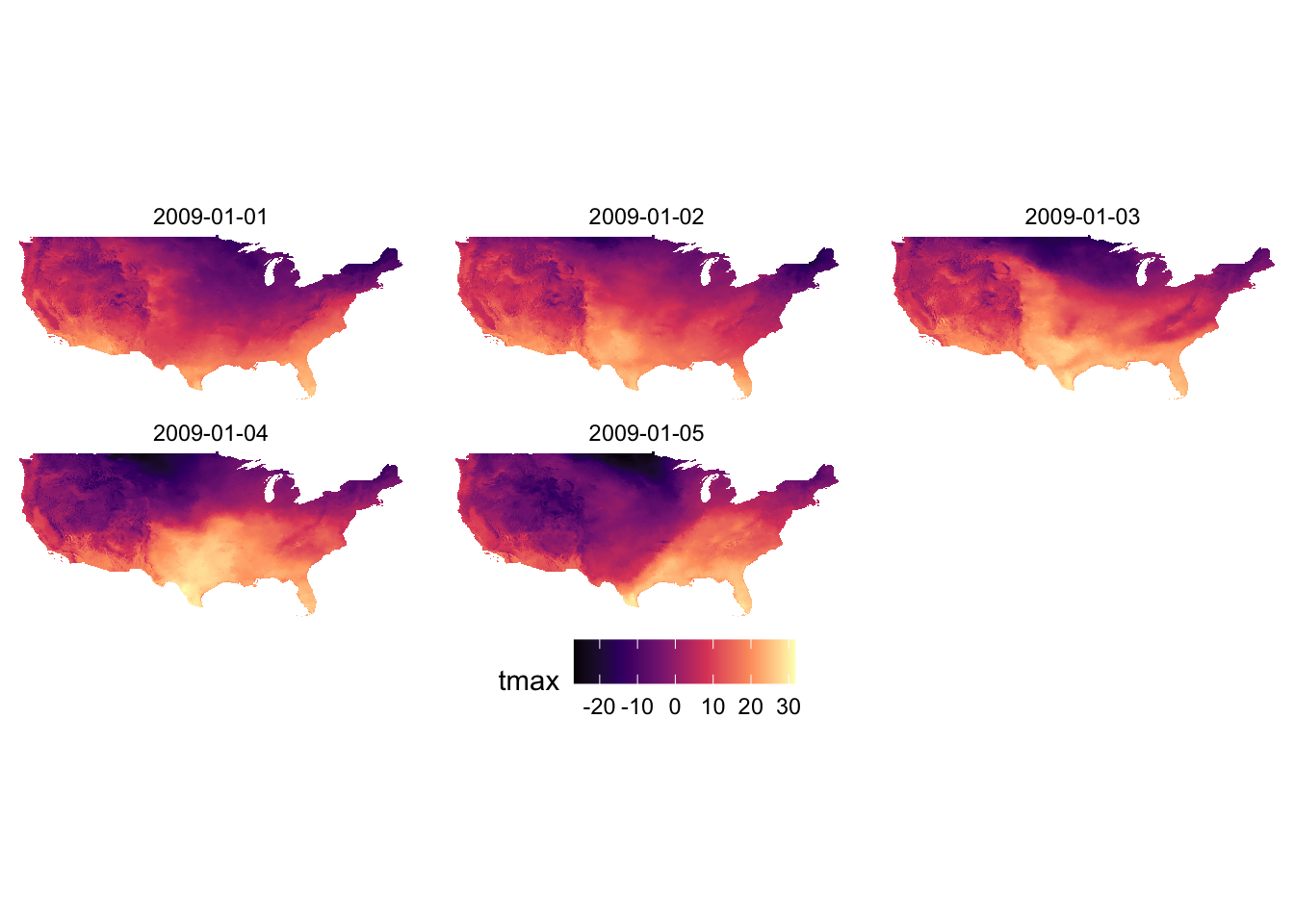
g_col_scale + scale_fill_viridis_c(option = "D")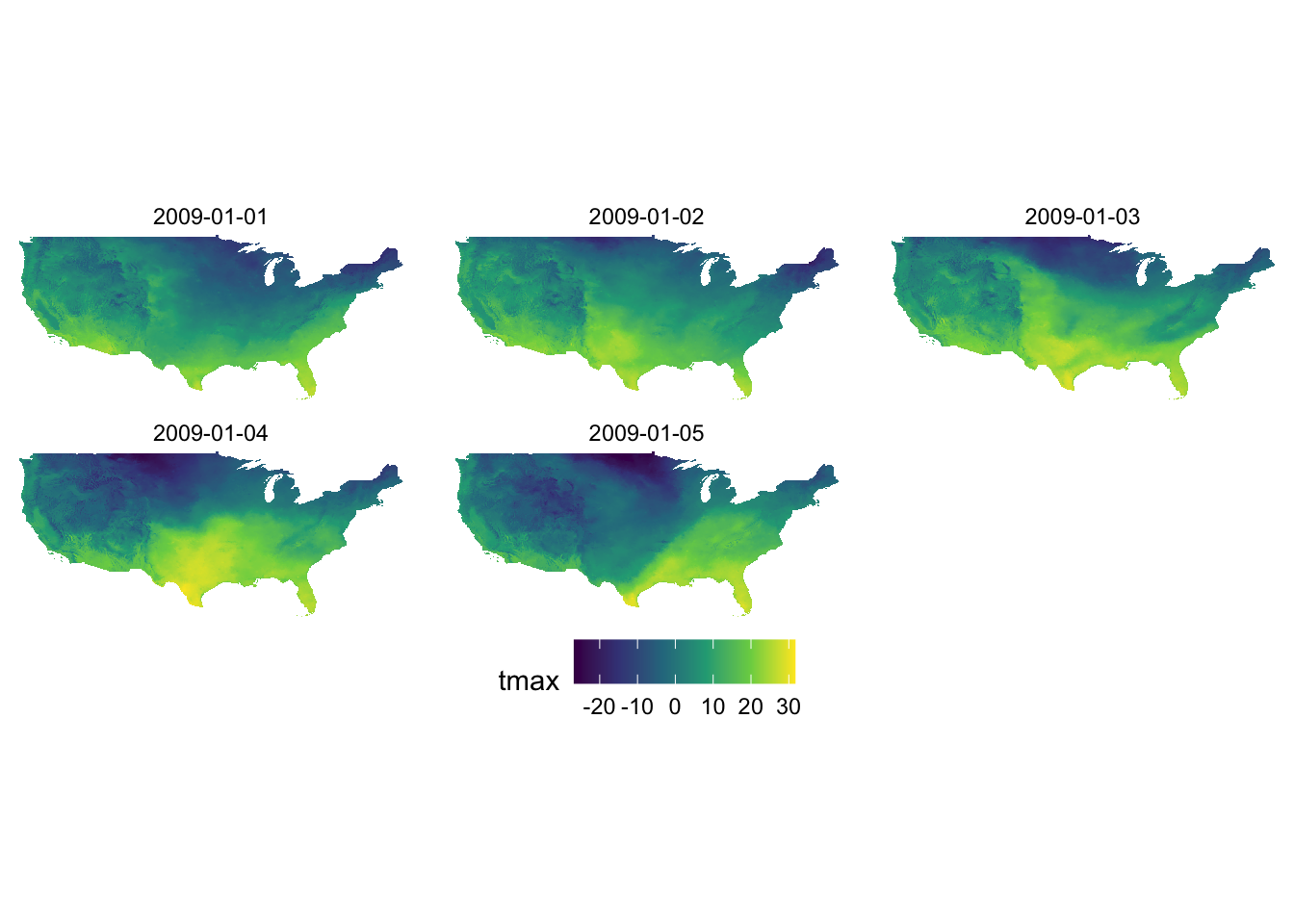
You can reverse the order of the color by adding direction = -1.
g_col_scale + scale_fill_viridis_c(option = "D", direction = -1)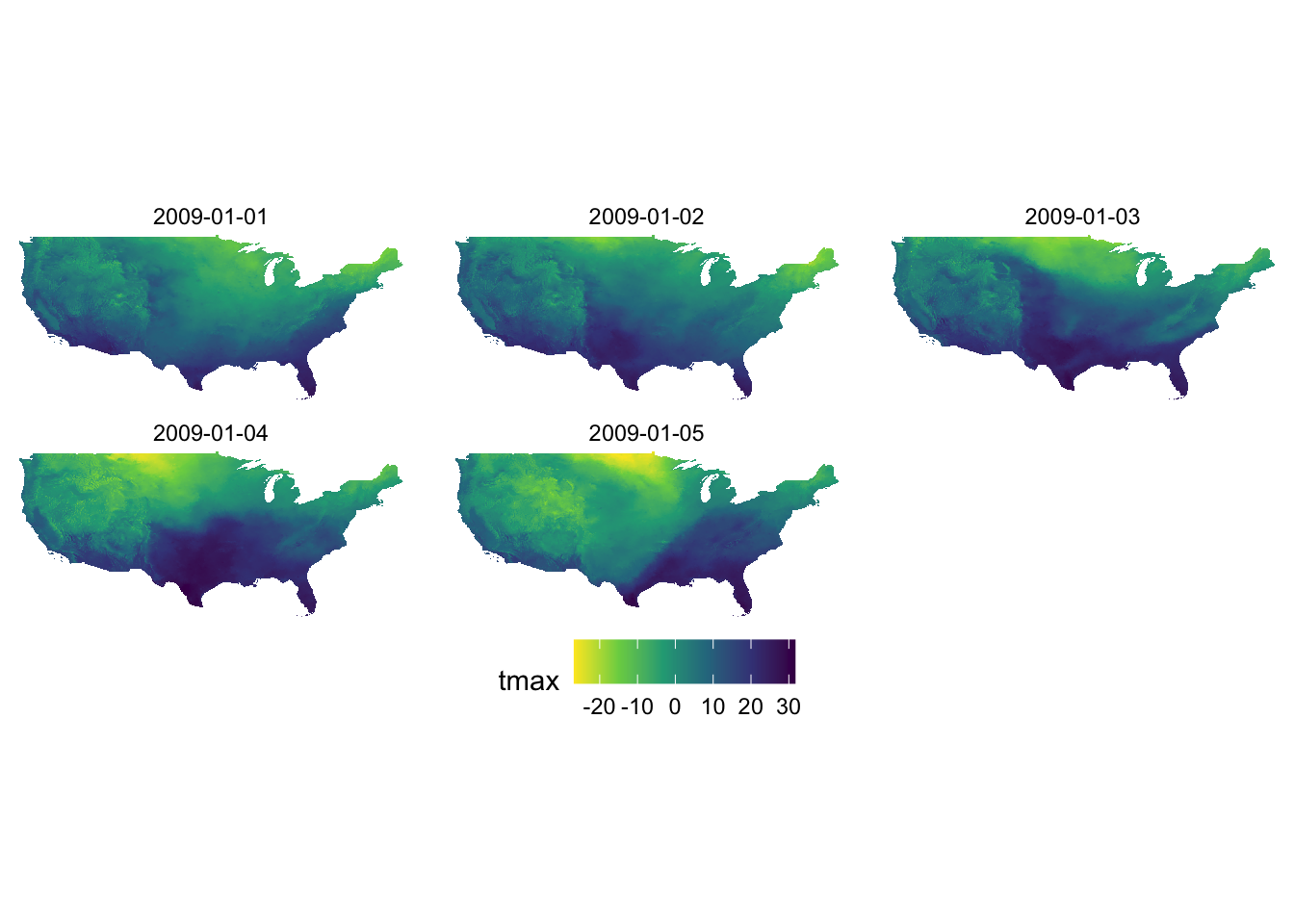
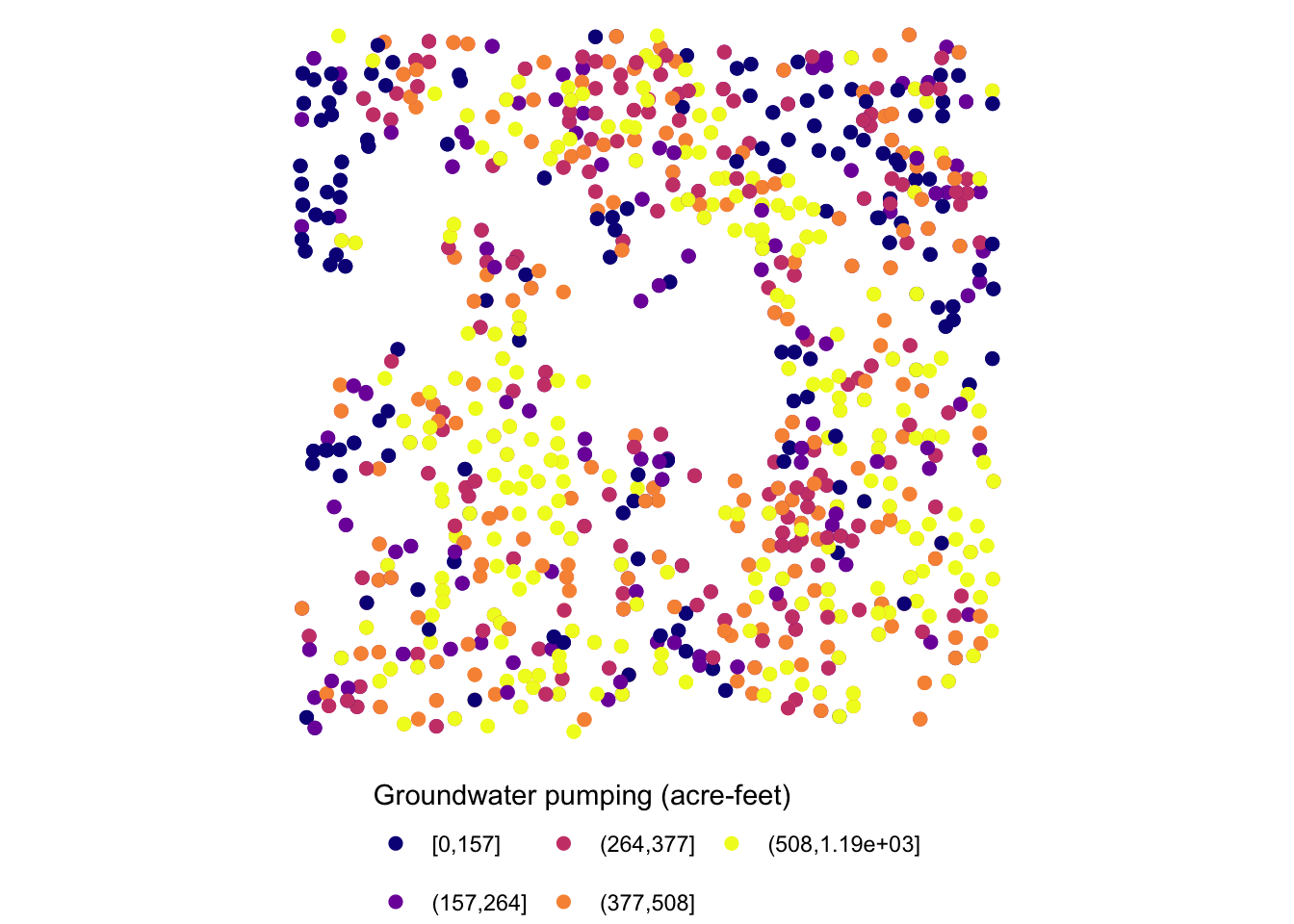
Let’s now work on aesthetics mapping based on a discrete variable. The code below groups af_used into five groups of ranges.
#--- convert af_used to a discrete variable ---#
gw_Stevens <- mutate(gw_Stevens, af_used_cat = cut_number(af_used, n = 5))
#--- take a look ---#
head(gw_Stevens)Simple feature collection with 6 features and 4 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -101.4232 ymin: 37.04683 xmax: -101.1111 ymax: 37.29295
Geodetic CRS: NAD83
well_id year af_used geometry af_used_cat
197 234 2010 461.9565 POINT (-101.4232 37.1165) (377,508]
198 234 2011 486.0534 POINT (-101.4232 37.1165) (377,508]
247 290 2010 457.4391 POINT (-101.2301 37.29295) (377,508]
248 290 2011 580.4156 POINT (-101.2301 37.29295) (508,1.19e+03]
275 317 2010 258.0000 POINT (-101.1111 37.04683) (157,264]
276 317 2011 255.0000 POINT (-101.1111 37.04683) (157,264]Since we would like to control color aesthetics based on a discrete variable, we should be using scale_color_viridis_d().
ggplot(data = gw_Stevens) +
geom_sf(aes(color = af_used_cat), size = 2) +
scale_color_viridis_d(option = "C")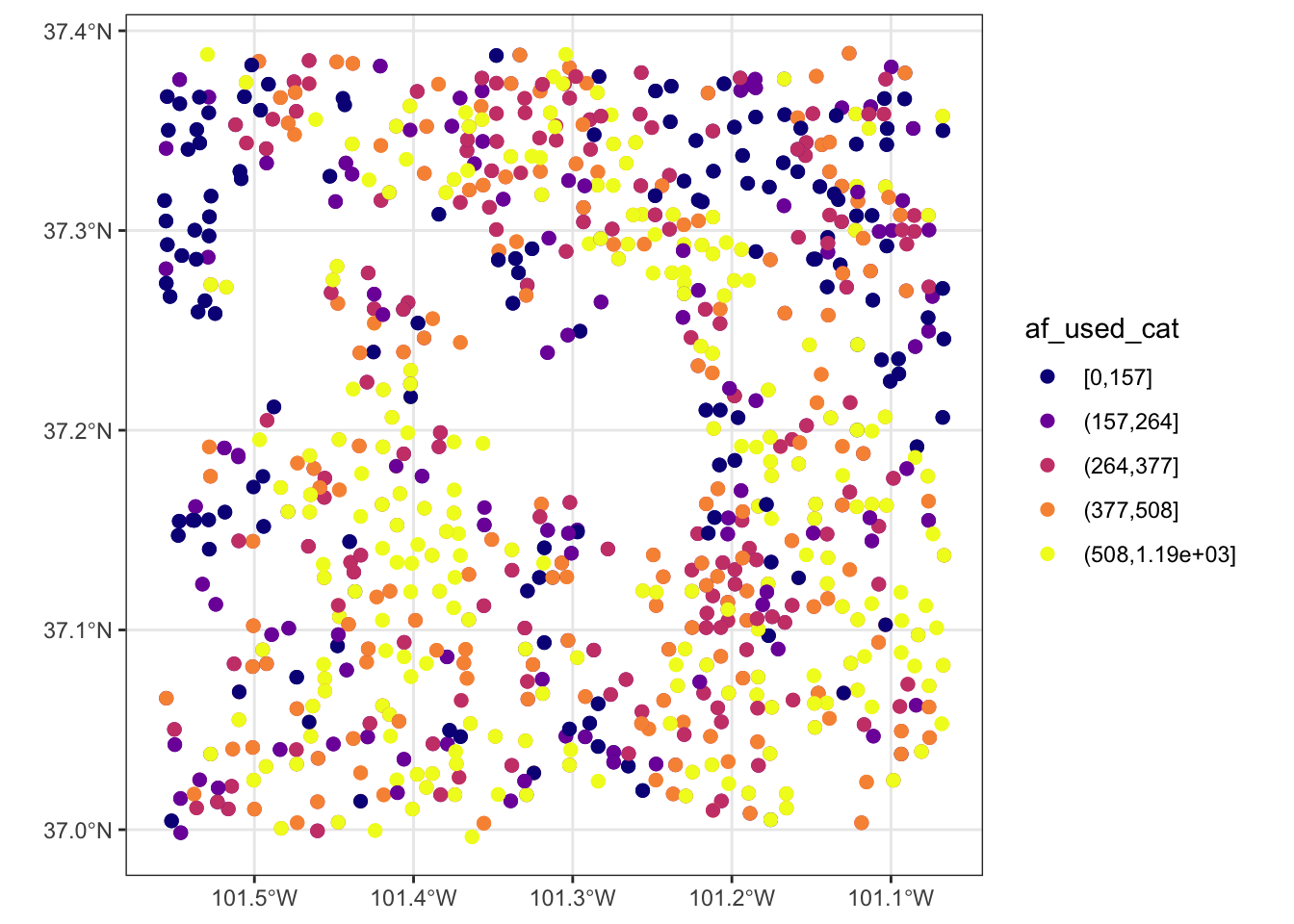
8.3.2 RColorBrewer: scale_*_distiller() and scale_*_brewer()
The RColorBrewer package provides a set of color scales that are useful. Here is the list of color scales you can use.
#--- load RColorBrewer ---#
library(RColorBrewer)
#--- disply all the color schemes from the package ---#
display.brewer.all()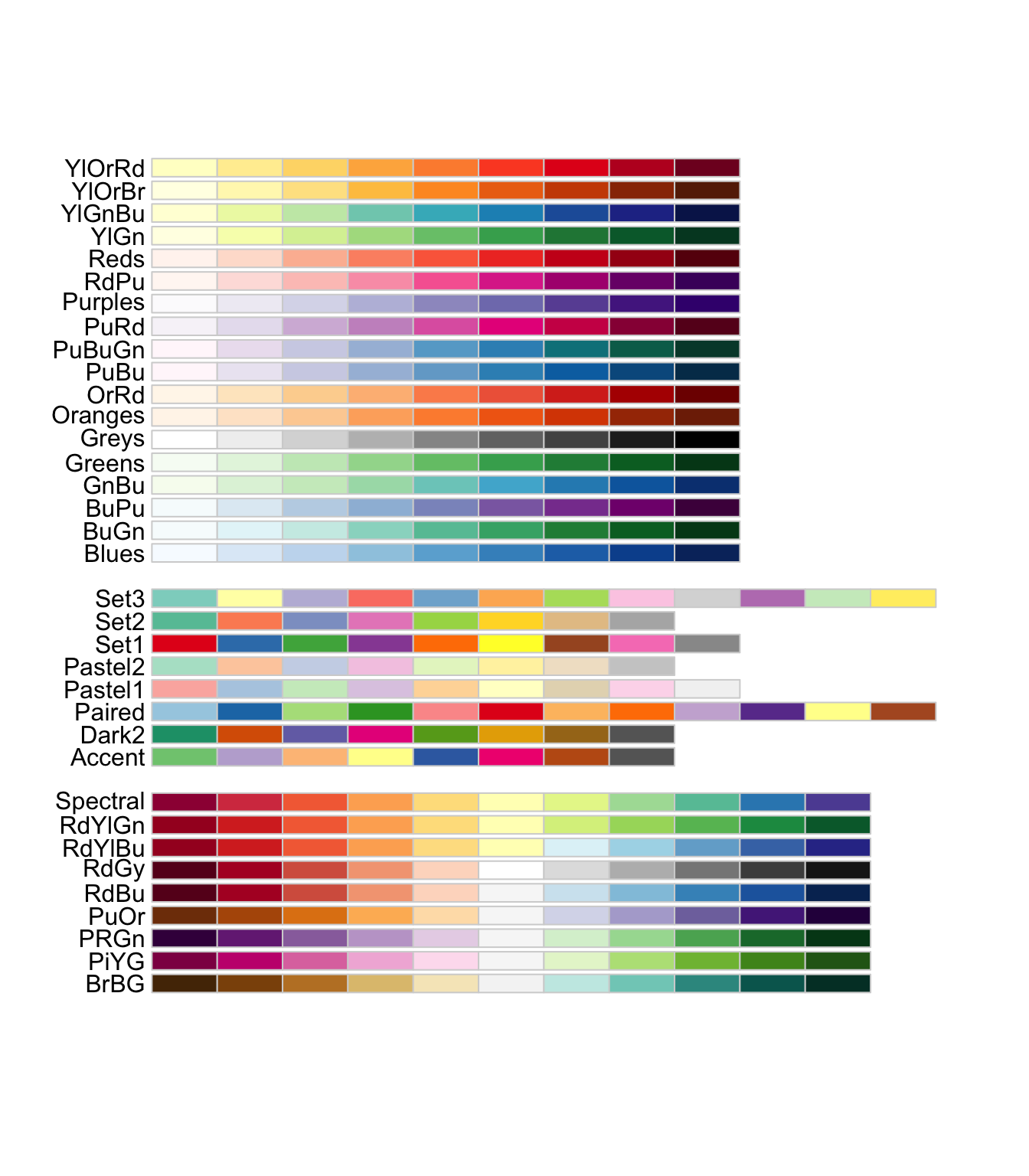
The first set of color palettes are sequential palettes and are suitable for a variable that has ordinal meaning: temperature, precipitation, etc. The second set of palettes are qualitative palettes and suitable for qualitative or categorical data. Finally, the third set of palettes are diverging palettes and can be suitable for variables that take both negative and positive values like changes in groundwater level.
Two types of scale functions can be used to use these palettes:
-
scale_*_distiller()for a continuous variable -
scale_*_brewer()for a discrete variable
To use a specific color palette, you can simply add palette = "palette name" inside scale_fill_distiller(). The codes below applies “Spectral” as an example.
g_col_scale + theme_void() +
scale_fill_distiller(palette = "Spectral")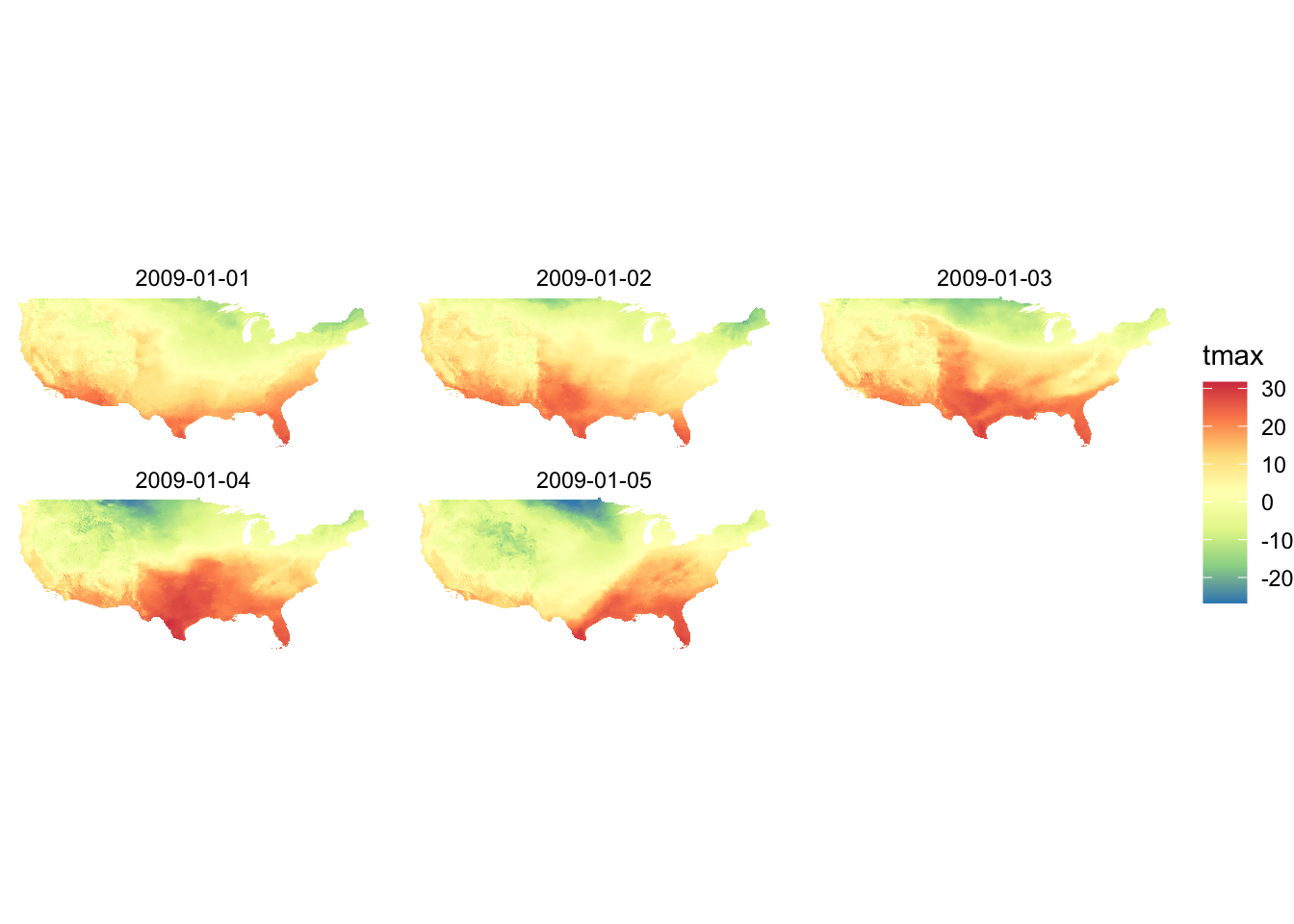
You can reverse the color order by adding trans = "reverse" option.
g_col_scale + theme_void() +
scale_fill_distiller(palette = "Spectral", trans = "reverse")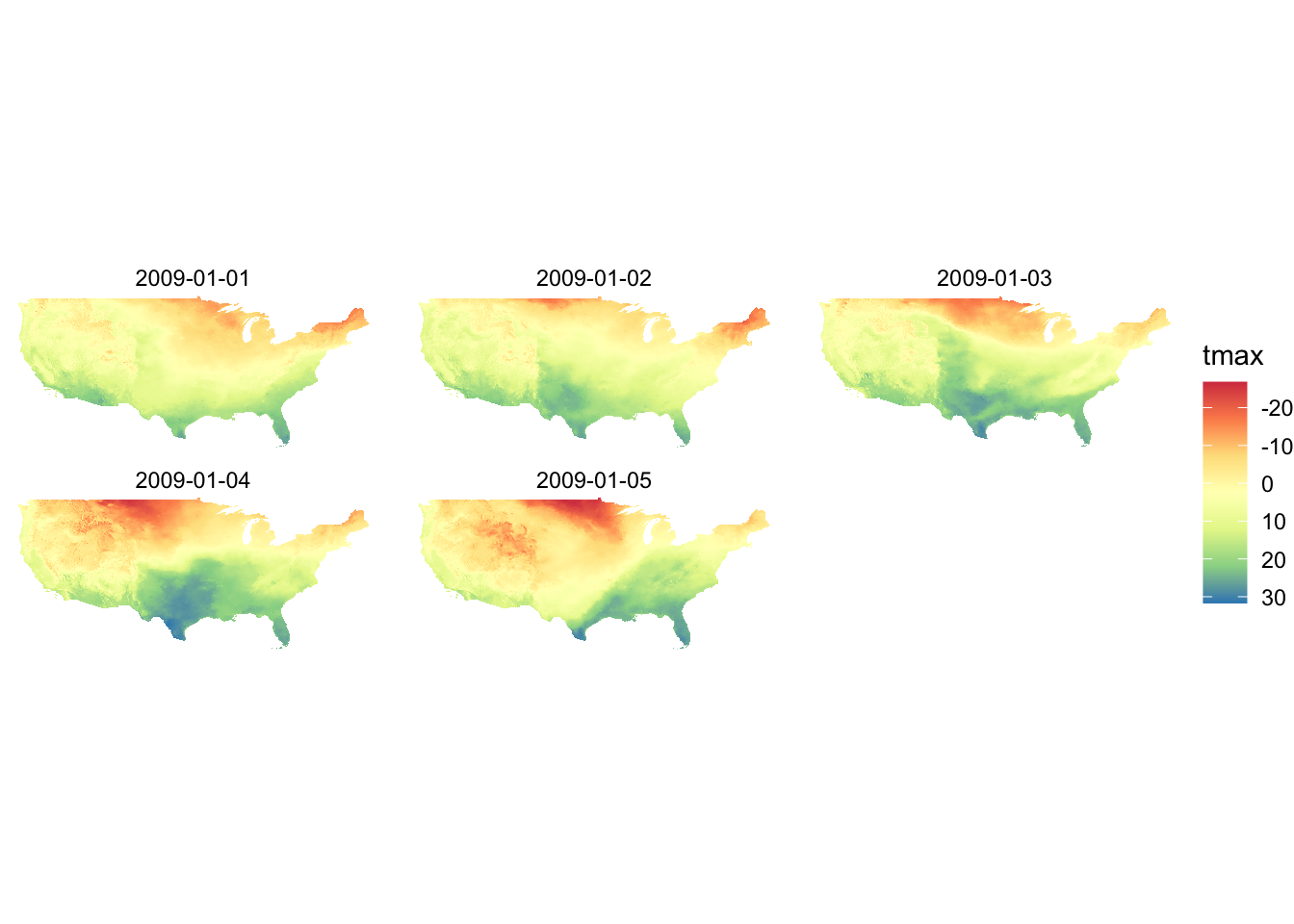
If you are specifying the color aesthetics based on a continuous variable, then you use scale_color_distiller().
ggplot(data = gw_Stevens) +
geom_sf(aes(color = af_used), size = 2) +
scale_color_distiller(palette = "Spectral")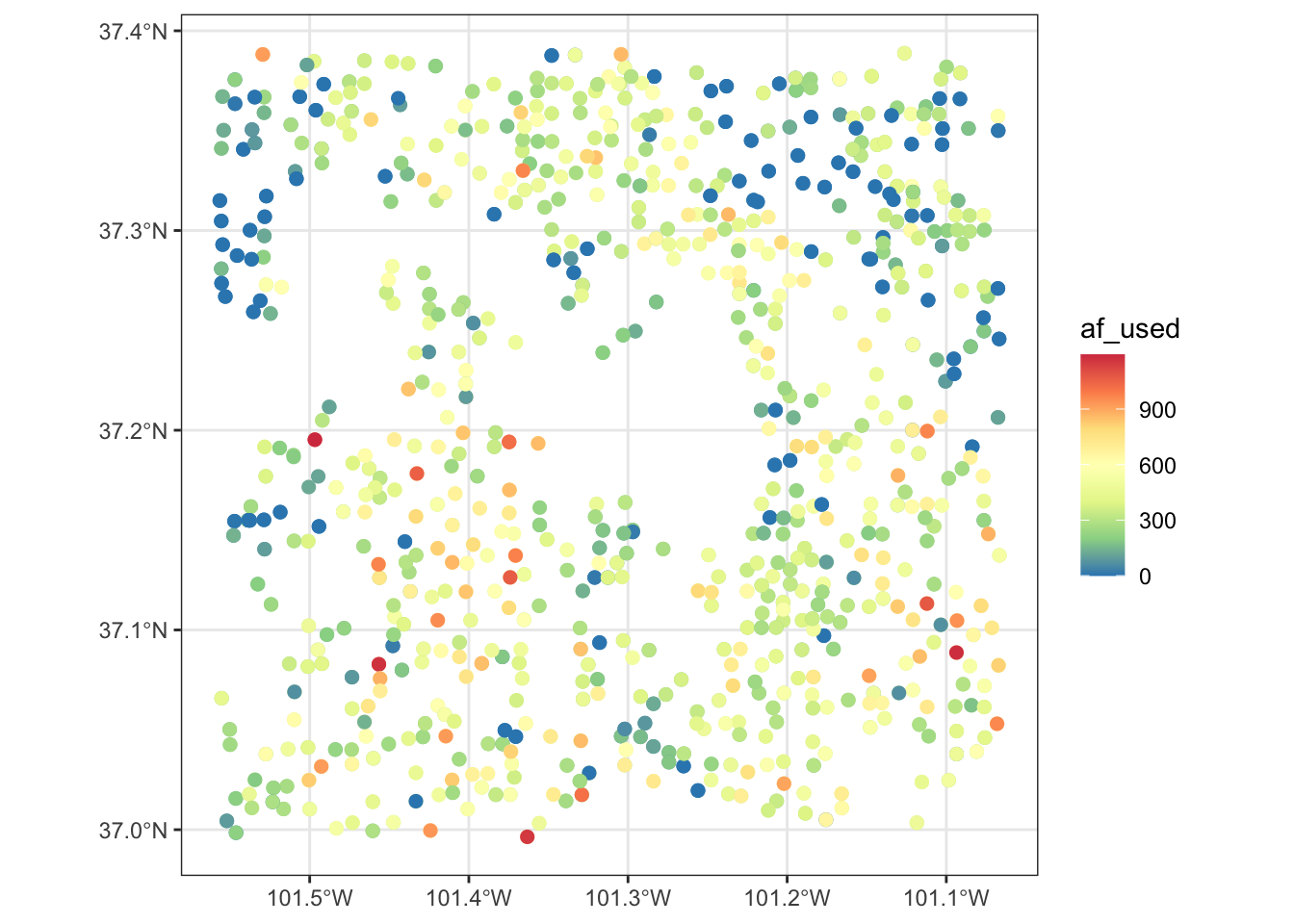
Now, suppose the variable of interest comes with categories of ranges of values. The code below groups af_used into five ranges using ggplo2::cut_number().
gw_Stevens <- mutate(gw_Stevens, af_used_cat = cut_number(af_used, n = 5))Since af_used_cat is a discrete variable, you can use scale_color_brewer() instead.
ggplot(data = gw_Stevens) +
geom_sf(aes(color = af_used_cat), size = 2) +
scale_color_brewer(palette = "Spectral")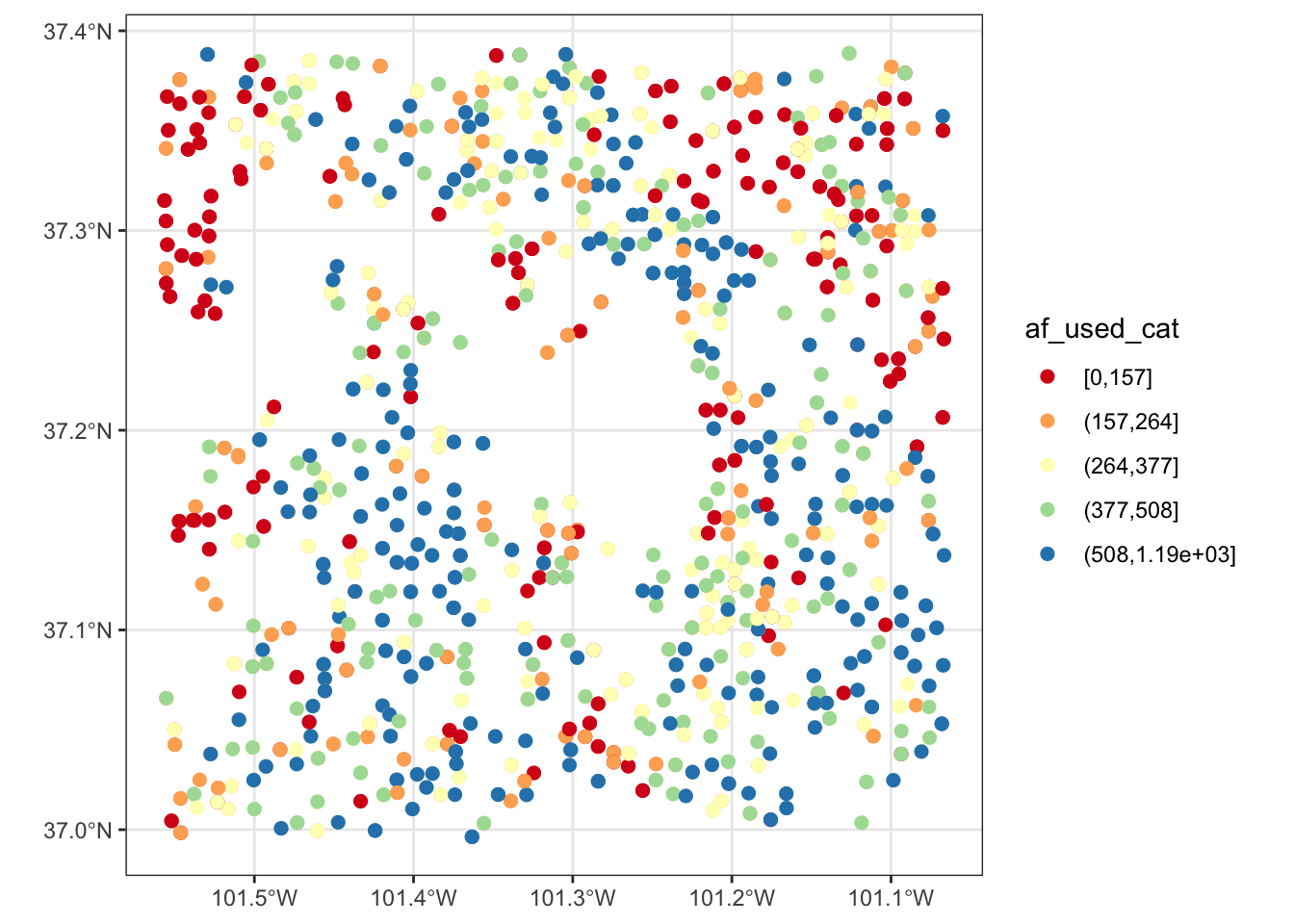
8.3.3 colorspace package
If you are not satisfied with the viridis color map or the ColorBrewer palette options, you might want to try the colorspace package.
Here is the palettes the colorspace package offers.
#--- plot the palettes ---#
hcl_palettes(plot = TRUE)
The packages offers its own scale_*() functions that follows the following naming convention:
scale_aesthetic_datatype_colorscale where
- aesthetic:
fillorcolor - datatype:
continuousordiscrete - colorscale:
qualitative,sequential,diverging,divergingx
For example, to add a sequential color scale to the following map, we would use scale_fill_continuous_sequential() and then pick a palette from the set of sequential palettes shown above. The code below uses the Viridis palette with the reverse option:
ggplot() +
geom_sf(data = gw_by_county, aes(fill = af_used)) +
facet_wrap(. ~ year) +
scale_fill_continuous_sequential(palette = "Viridis", trans = "reverse")If you still cannot find a palette that satisfies your need (or obsession at this point), then you can easily make your own. The package offers hclwizard(), which starts shiny-based web application to let you design your own color palette.
After running this,
hclwizard()you should see a web application pop up that looks like this.
knitr::include_graphics("hclwizard.png")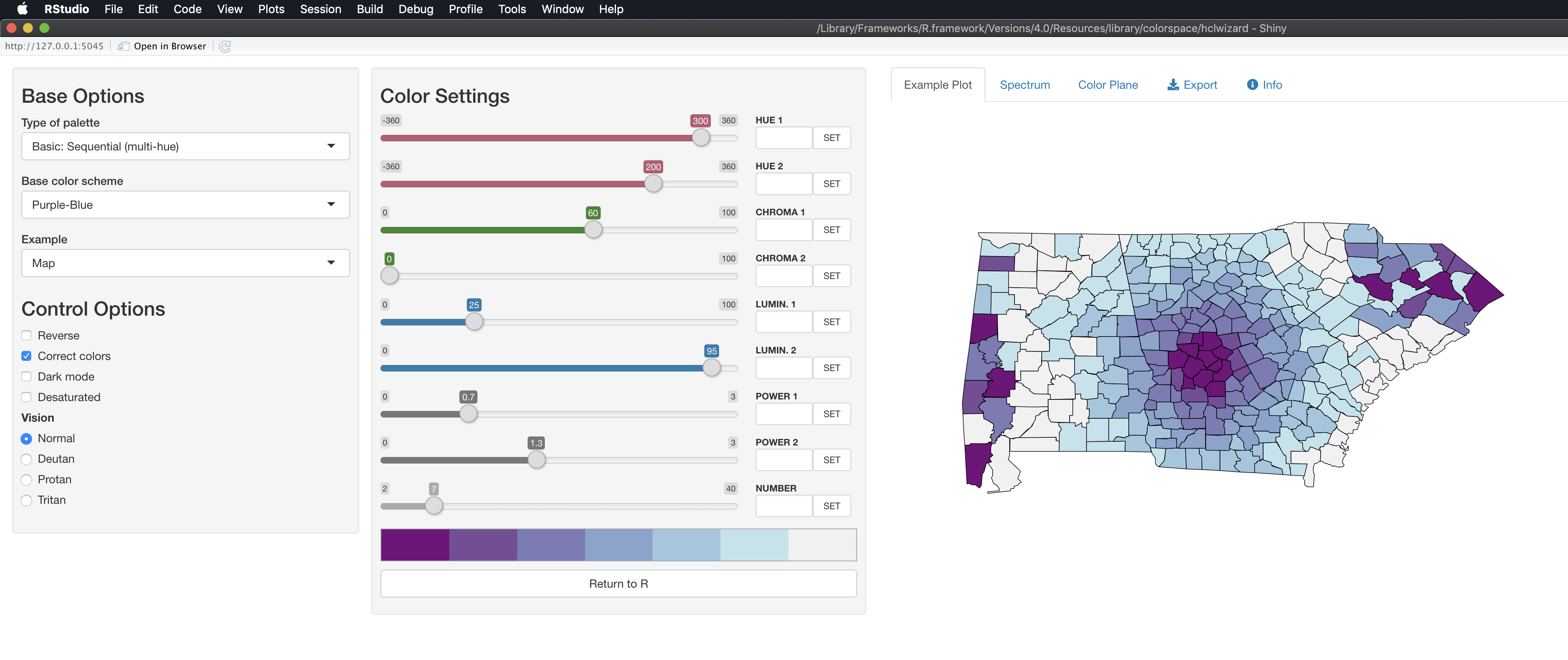
After you find a color scale you would like to use, you can go to the Exporttab, select the R tab, and then copy the code that appear in the highlighted area.
knitr::include_graphics("hclwizard_pick_theme.png")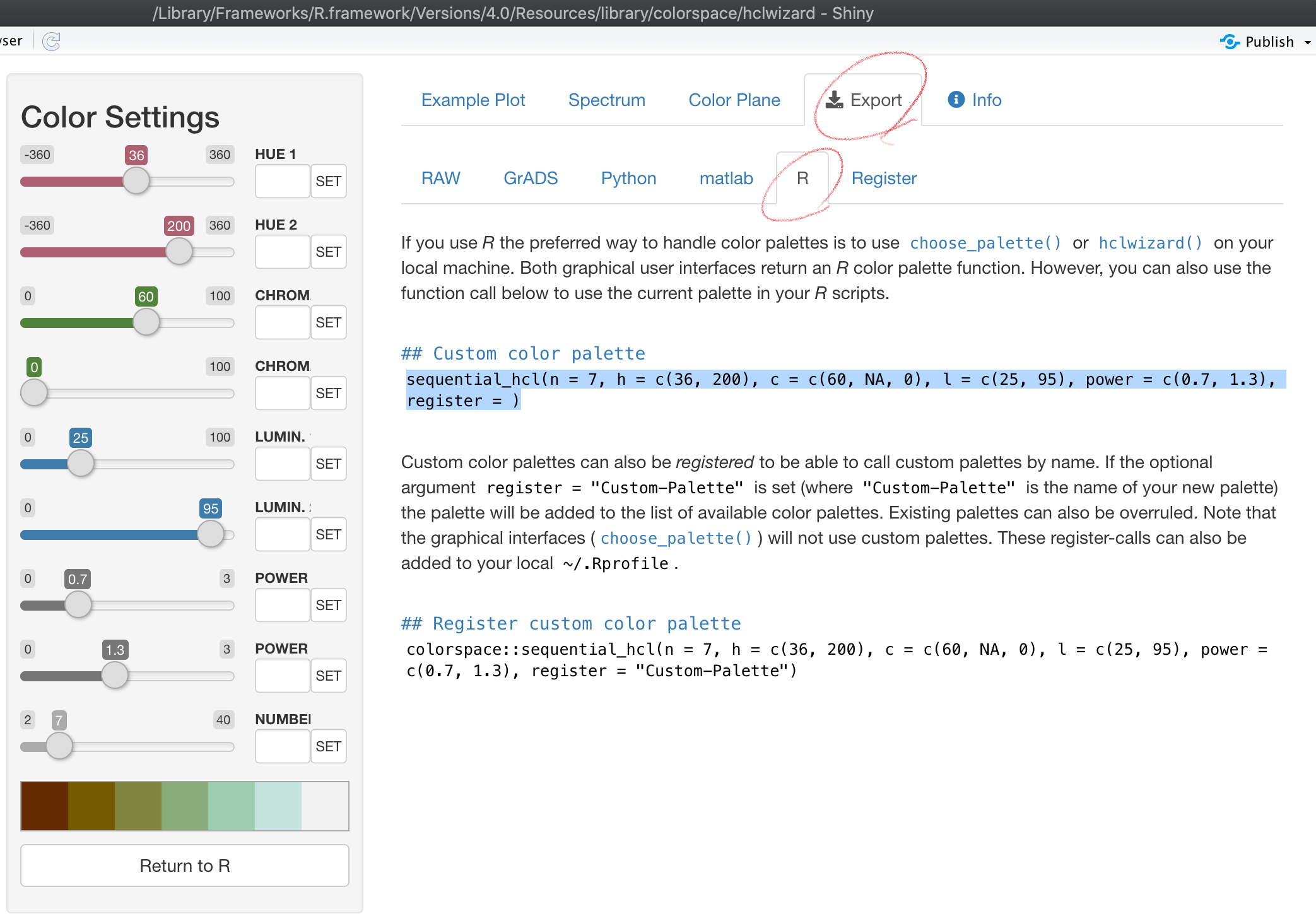
You could register the color palette by completing the register = option in the copied code if you think you will use it other times. Otherwise, you can delete the option.
col_palette <- sequential_hcl(n = 7, h = c(36, 200), c = c(60, NA, 0), l = c(25, 95), power = c(0.7, 1.3))We then use the code as follows:
g_col_scale + theme_void() +
scale_fill_gradientn(colors = col_palette)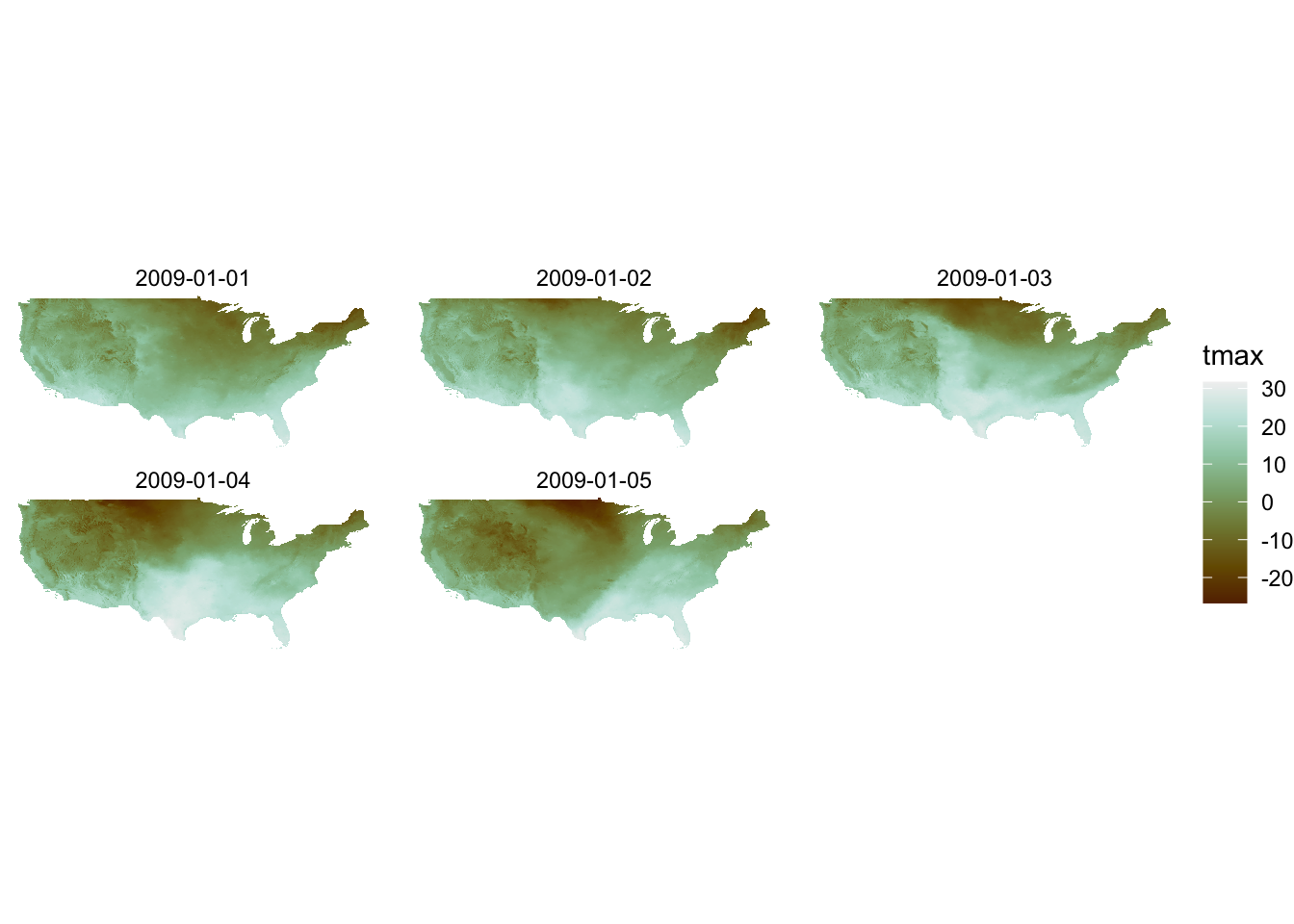
Note that you are now using scale_*_gradientn() with this approach.
For a discrete variable, you can use scale_*_manual():
col_discrete <- sequential_hcl(n = 5, h = c(240, 130), c = c(30, NA, 33), l = c(25, 95), power = c(1, NA), rev = TRUE)
ggplot() +
geom_sf(data = gw_Stevens, aes(color = af_used_cat), size = 2) +
scale_color_manual(values = col_discrete)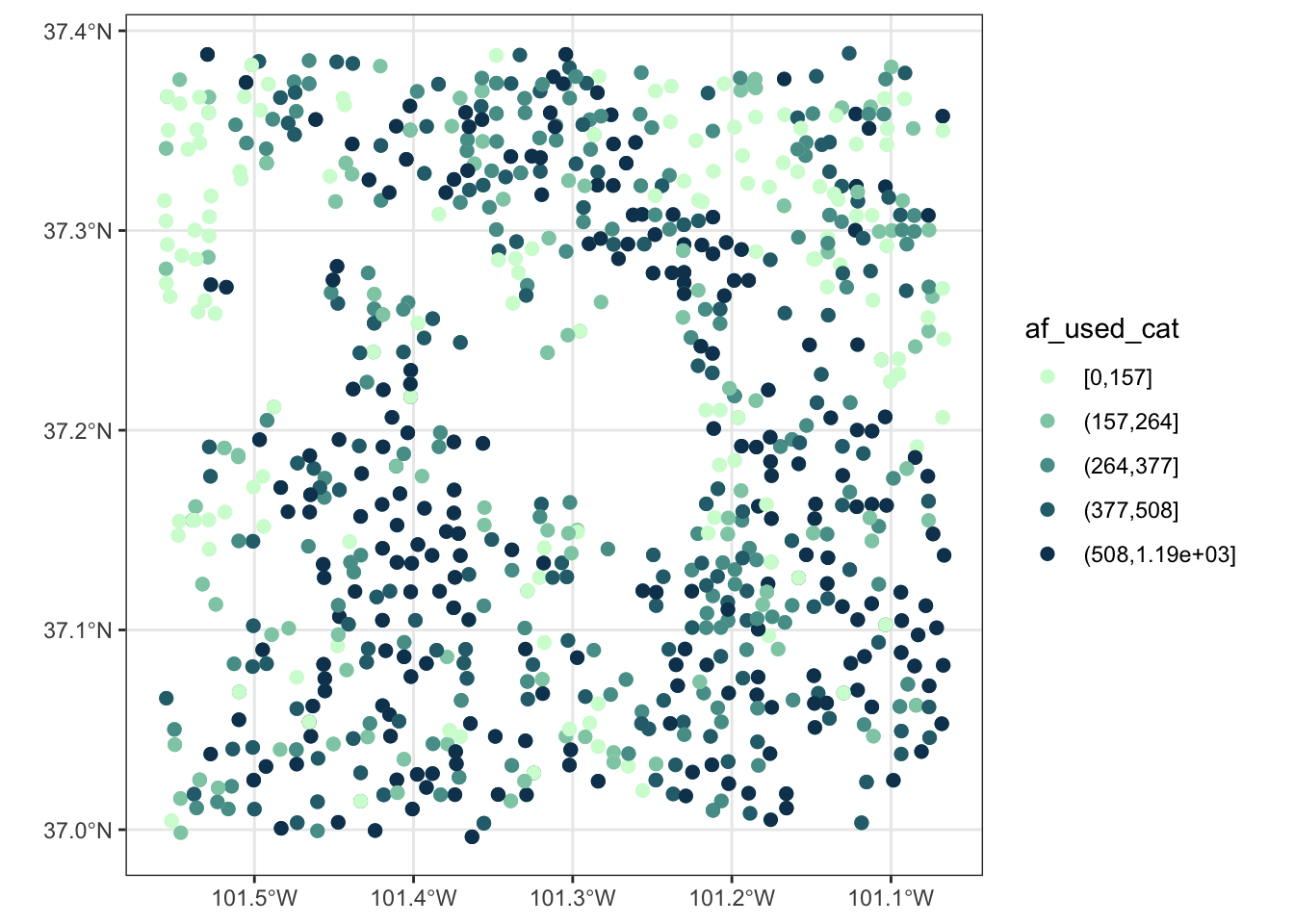
8.4 Arranging maps
8.4.1 Multiple panels of figures as a single figure
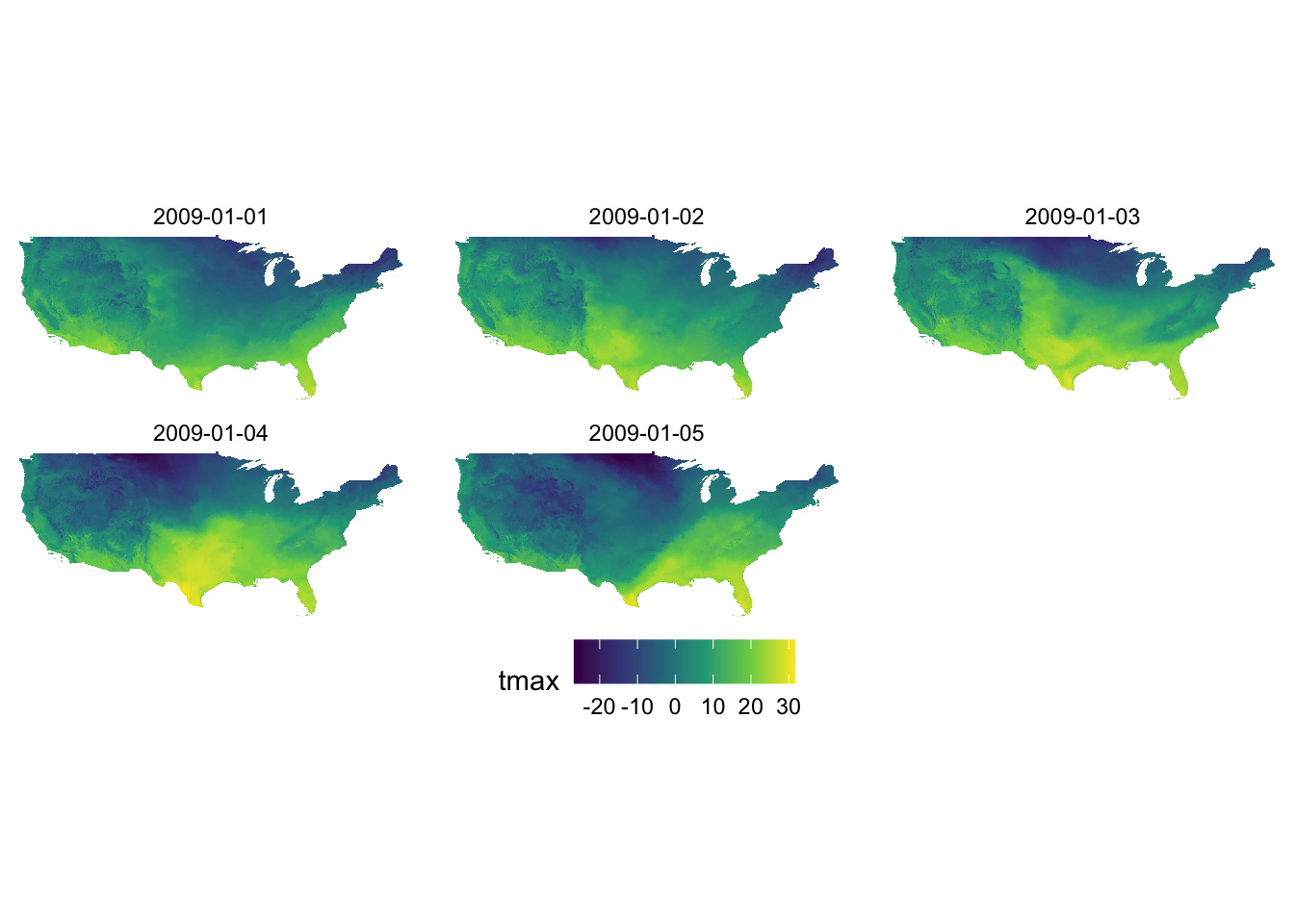
Faceting using facet_wrap() or facet_grid() allows for dividing the data into groups and creating a map for each group. It is particularly suitable for visualizing one variable at different facets. A good example is a collection of maps of tmax observed at different dates (see figure below). Faceting provides a single consistent color scale shared across the facets.
ggplot() +
geom_raster(data = tmax_long_df, aes(x = x, y = y, fill = tmax)) +
facet_wrap(date ~ .) +
coord_equal() +
scale_fill_viridis_c() +
theme_void() +
theme(
legend.position = "bottom"
)
However, faceting is not suitable for creating maps of different variables. To see this let’s plot tmax and precipitation on Jan 1, 2009 together. We first read precipitation data for Jan 1, 2009.
ppt_long_df <- readRDS("Data/ppt_long_df.rds")Let’s extract tmax and precipitation on Jan 1, 2009 from their respective datasets, and combine them.
#--- get tmax on Jan 1, 2009 ---#
tmax_df <- filter(tmax_long_df, date == ymd("2009-01-01")) %>%
setnames("tmax", "value") %>%
mutate(type = "tmax")
#--- get precipitation on Jan 1, 2009 ---#
ppt_df <- filter(ppt_long_df, date == ymd("2009-01-01")) %>%
mutate(type = "ppt")
#--- combine them ---#
combined_df <- rbind(tmax_df, ppt_df)Here is the map faceted for tmax and precipitation:
ggplot() +
geom_raster(data = combined_df, aes(x = x, y = y, fill = value)) +
facet_grid(type ~ .) +
scale_fill_viridis() +
theme_void()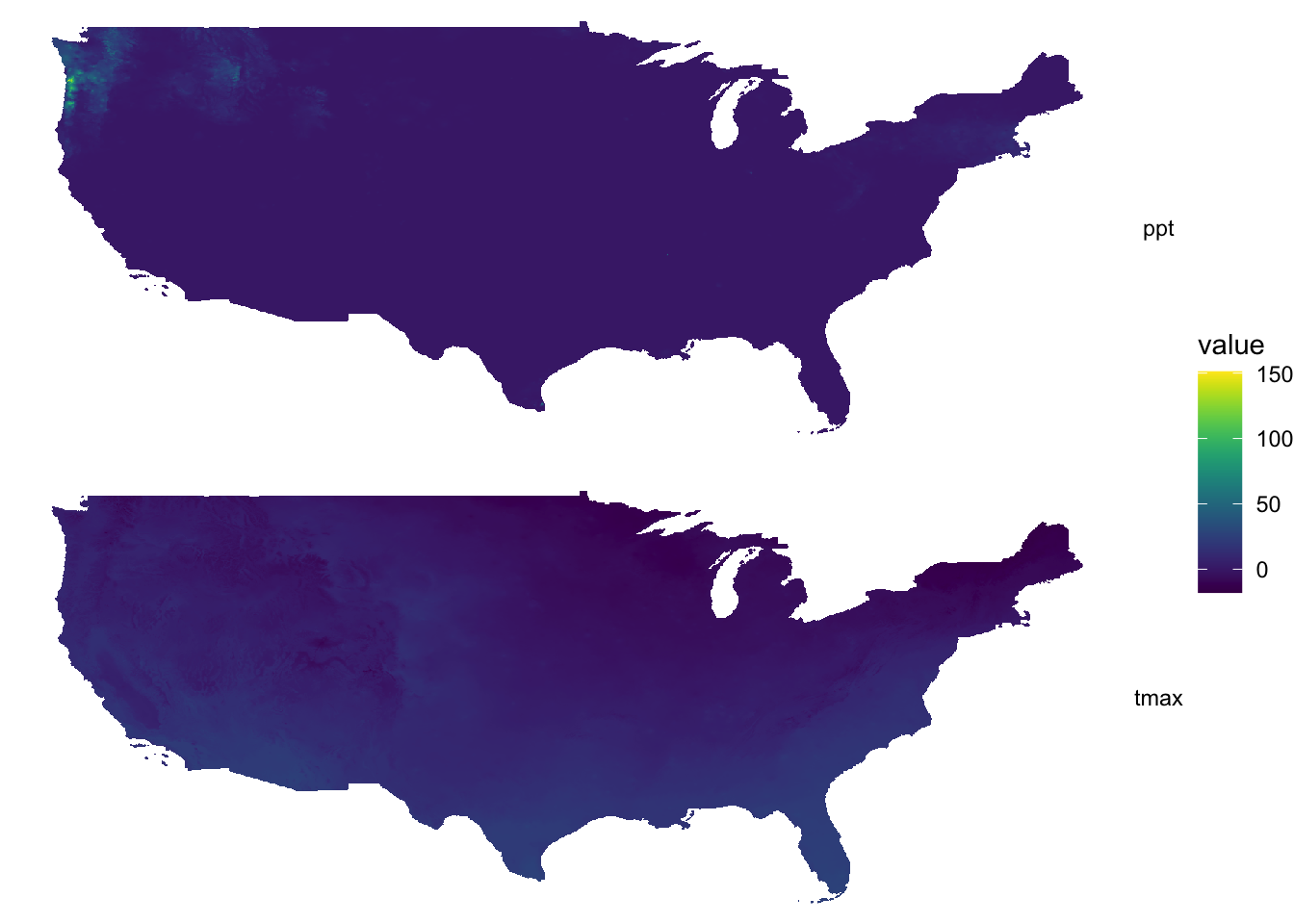
As you can see, a single color scale is created for precipitation recorded in mm and temperature observed in Celsius. On this particular day, precipitation of more than 150 mm was observed on a part of the west coast. Consequently, you see almost no color differentiation on the tmax map which ranges roughly from -20 to 30. It is simply not a good idea to facet for two variables observed at different scales.
Instead, you should have an independent color scale for each of the variables and then just combine them. Now, you might ask if you really need to combine the two. Can’t you just have two figures and arrange them in the manner you would like on a pdf or WORD document? If you are still convinced that you need to have two panels of figures as one figure, then you can use the patchwork package.
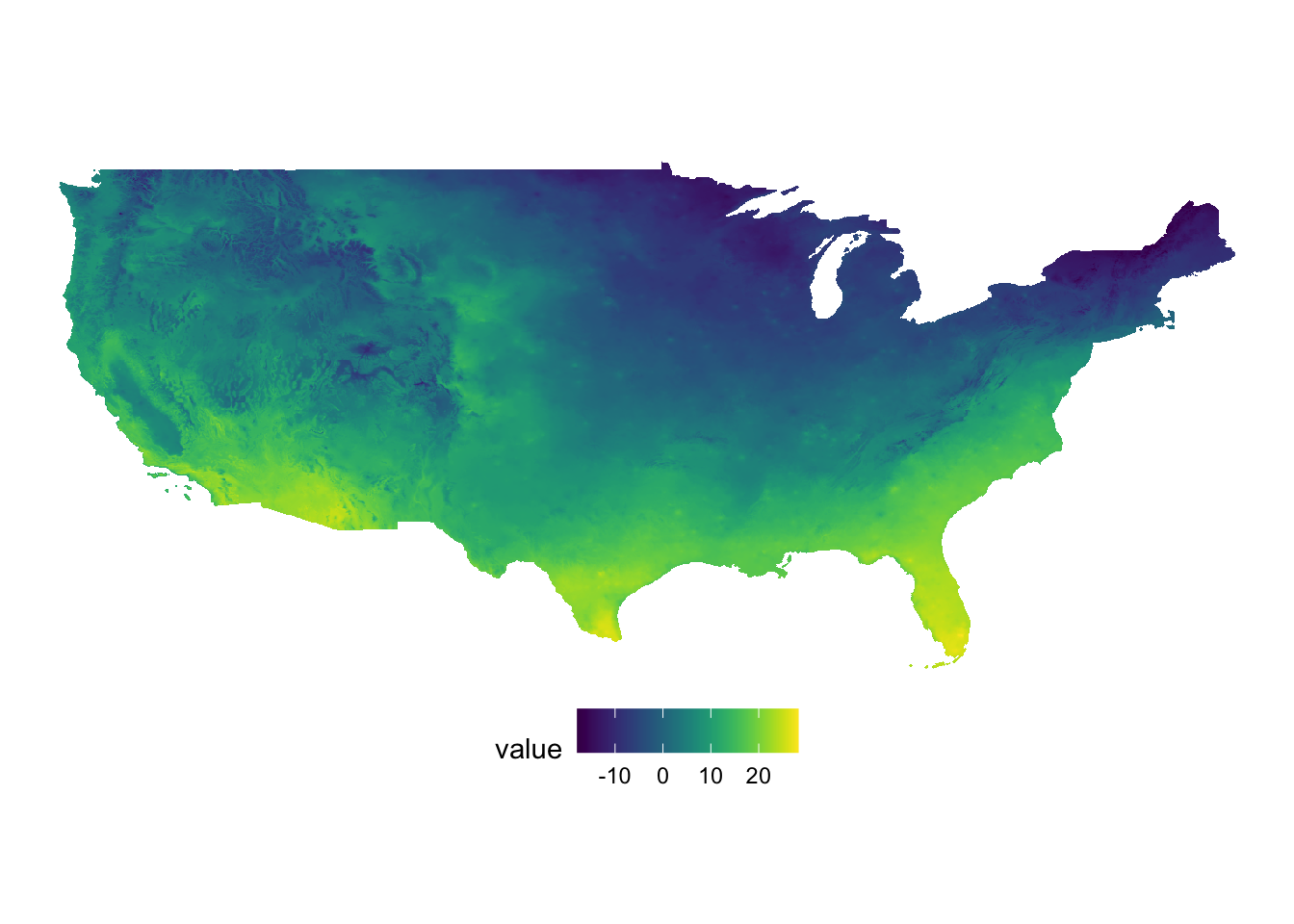
patchwork combines ggplot objects (maps) using simple operators: +, /, and |. Let’s first create maps of tmax and precipitation separately.
#--- tmax ---#
(
g_tmax <- ggplot() +
geom_raster(data = tmax_df, aes(x = x, y = y, fill = value)) +
scale_fill_viridis() +
theme_void() +
coord_equal() +
theme(legend.position = "bottom")
)
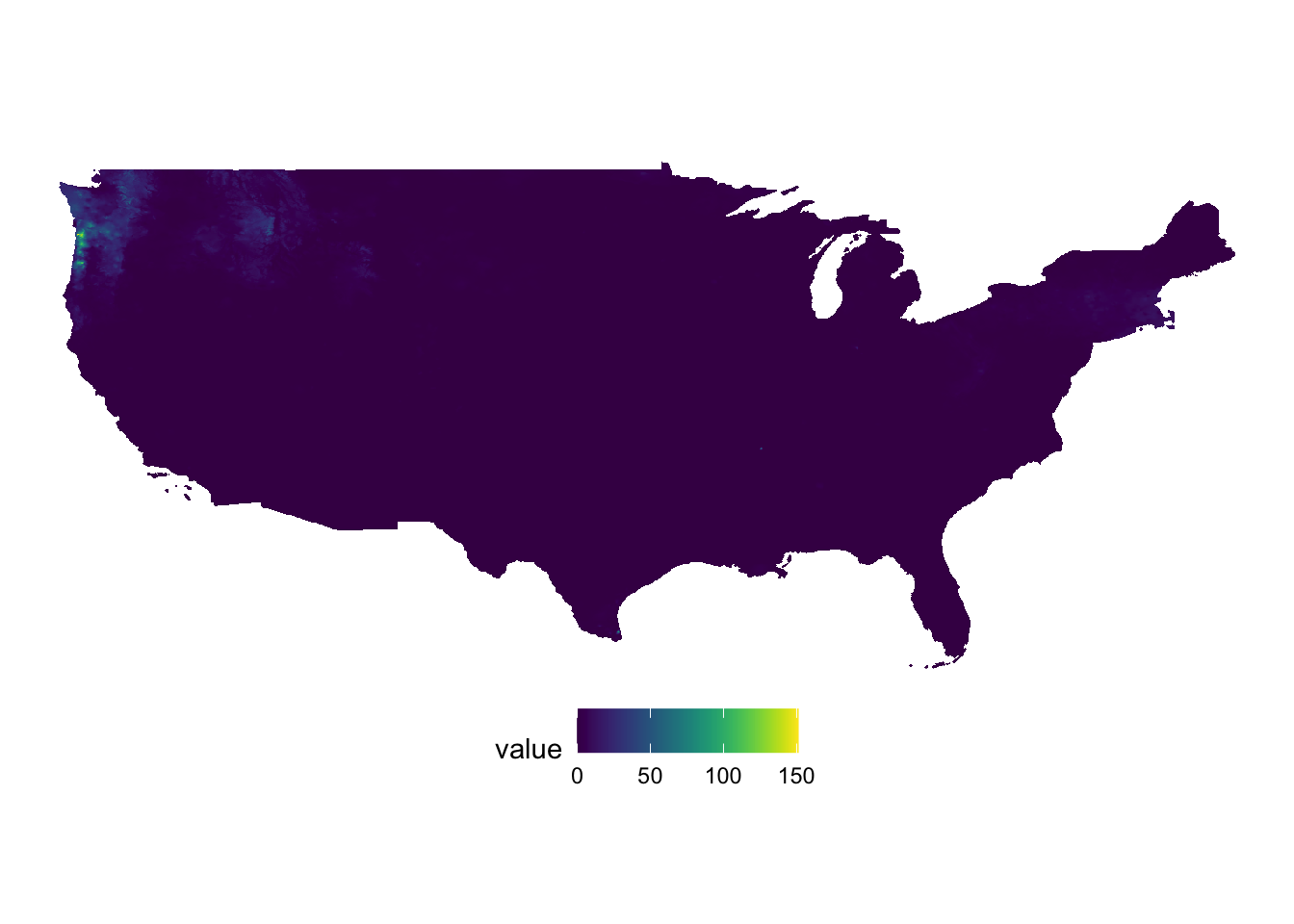
#--- ppt ---#
(
g_ppt <- ggplot() +
geom_raster(data = ppt_df, aes(x = x, y = y, fill = value)) +
scale_fill_viridis() +
theme_void() +
coord_equal() +
theme(legend.position = "bottom")
)
It is best to just look at examples to get the sense of how patchwork works. A fuller treatment of patchwork is found here (https://patchwork.data-imaginist.com/index.html).
Example 1
g_tmax + g_ppt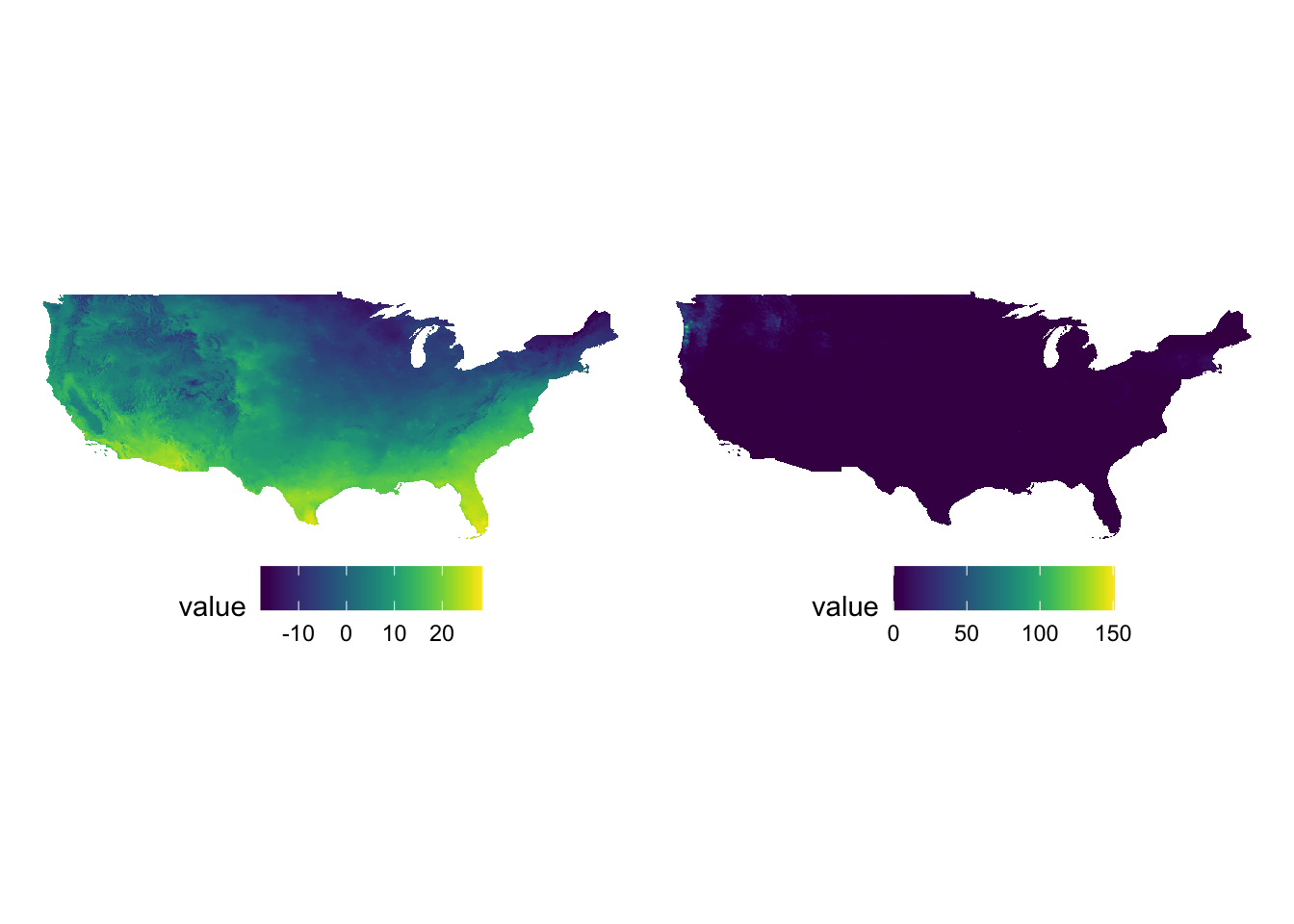
Example 2
g_tmax / g_ppt / g_tmax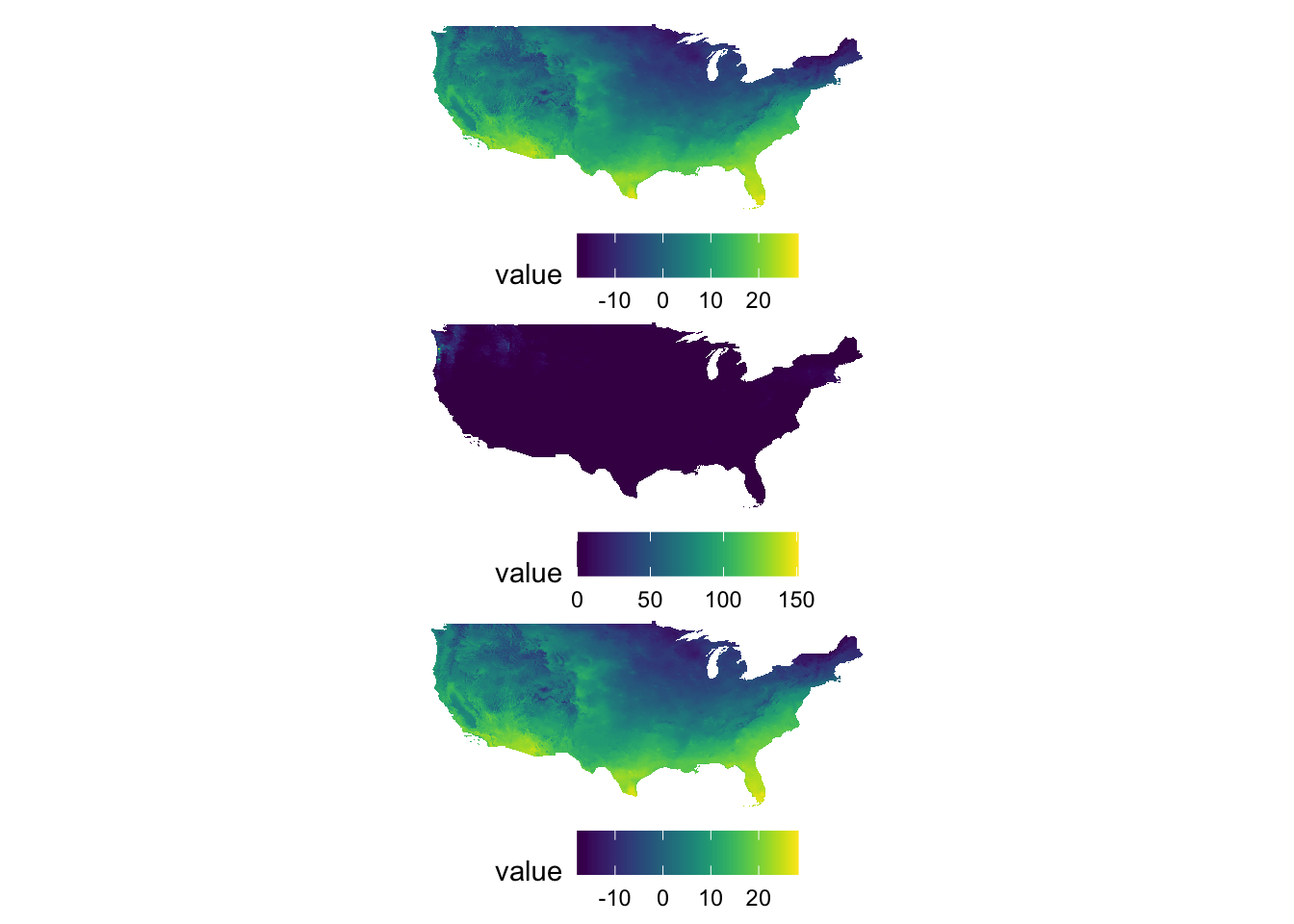
Example 3
g_tmax + g_ppt + plot_layout(nrow = 2)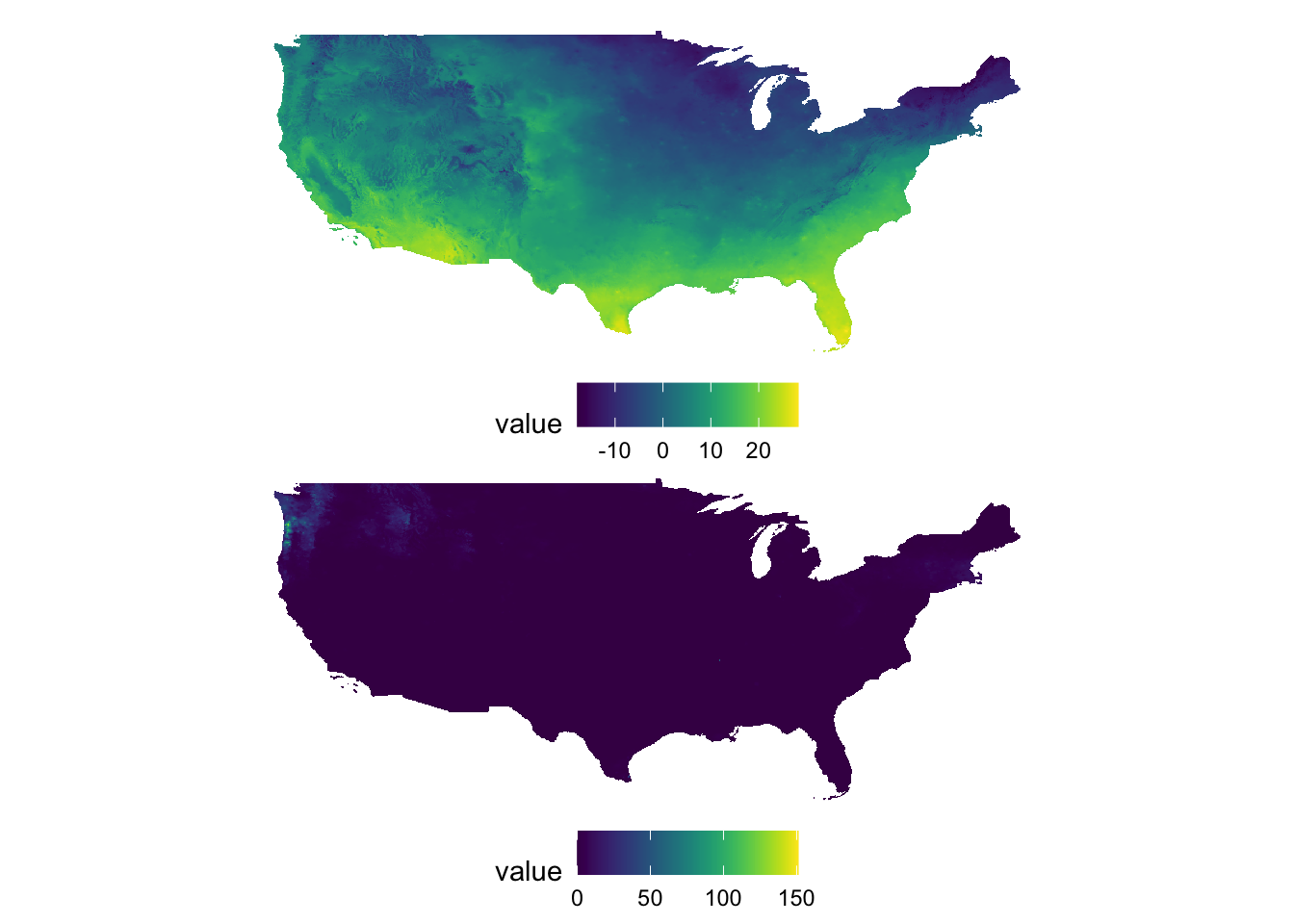
Example 4
g_tmax + g_ppt + g_tmax + g_ppt + plot_layout(nrow = 3, byrow = FALSE)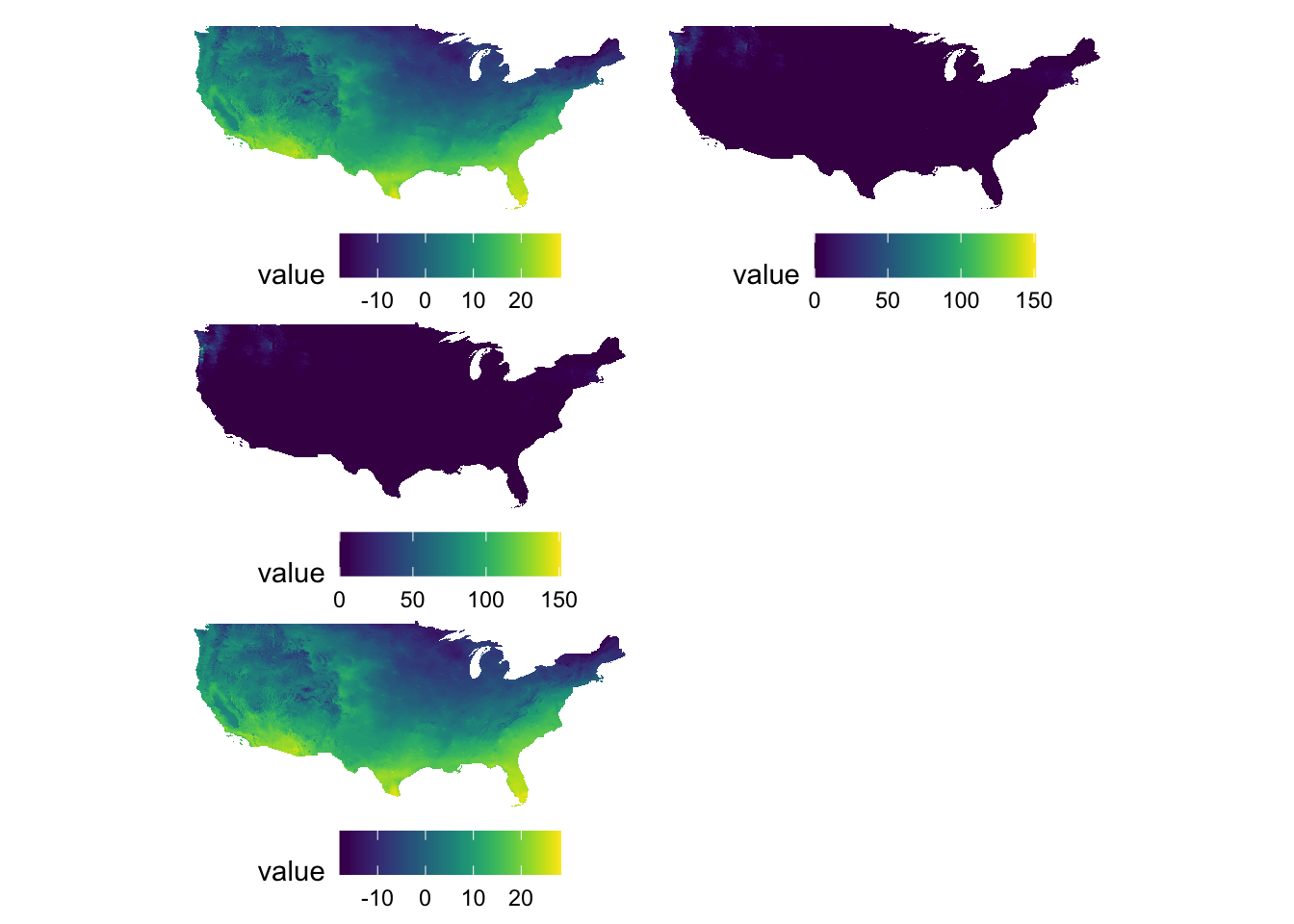
Example 5
g_tmax | (g_ppt / g_tmax)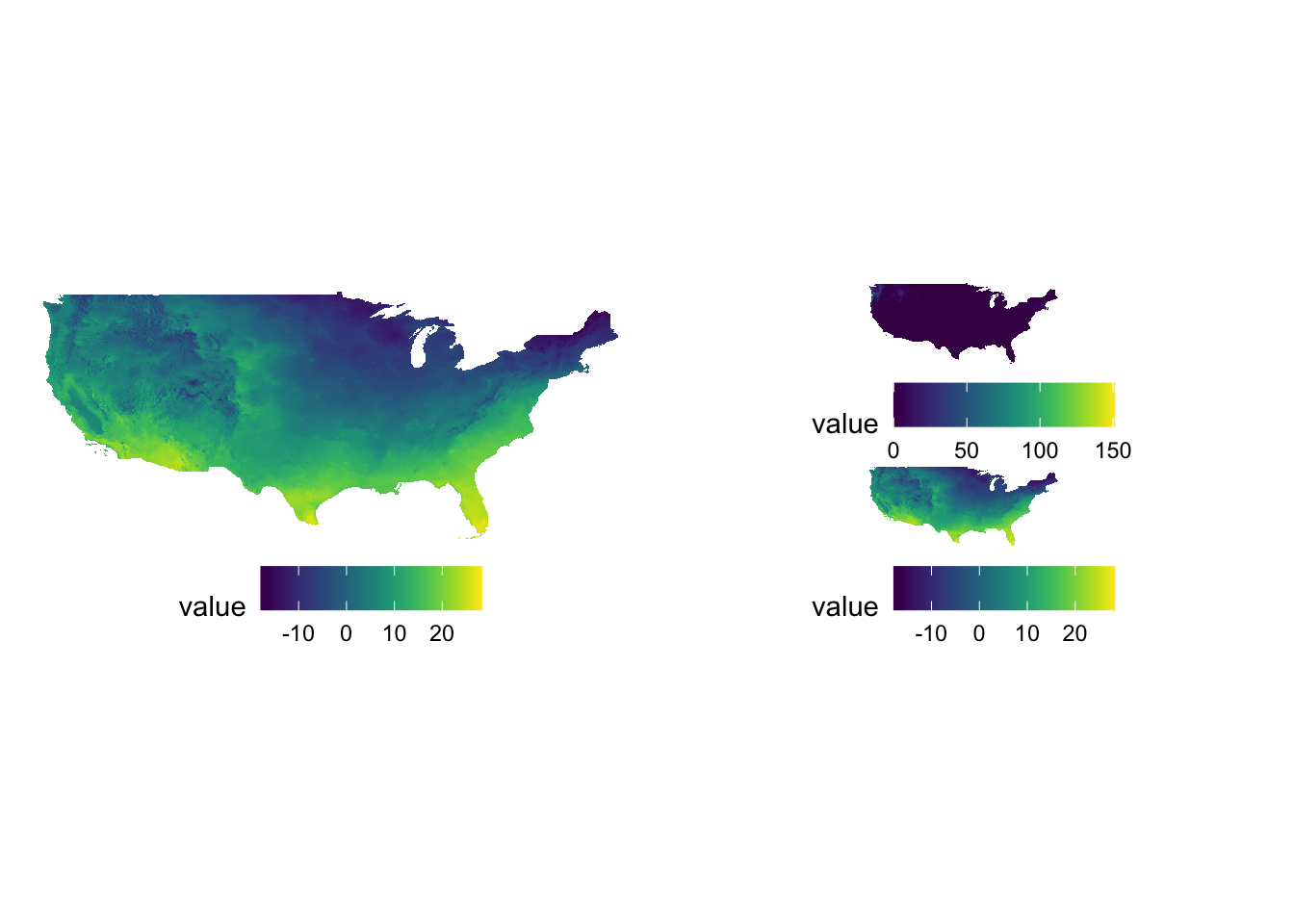
Sometimes figures are placed too close to each other. In such a case, you can pad a figure at the time of generating individual figures by adding the plot.margin option to theme(). For example, the following code creates space at the bottom of g_tmax (5 cm), and vertically stack g_tmax and g_ppt.
#--- space at the bottom ---#
g_tmax <- g_tmax + theme(plot.margin = unit(c(0, 0, 5, 0), "cm"))
#--- vertically stack ---#
g_tmax / g_ppt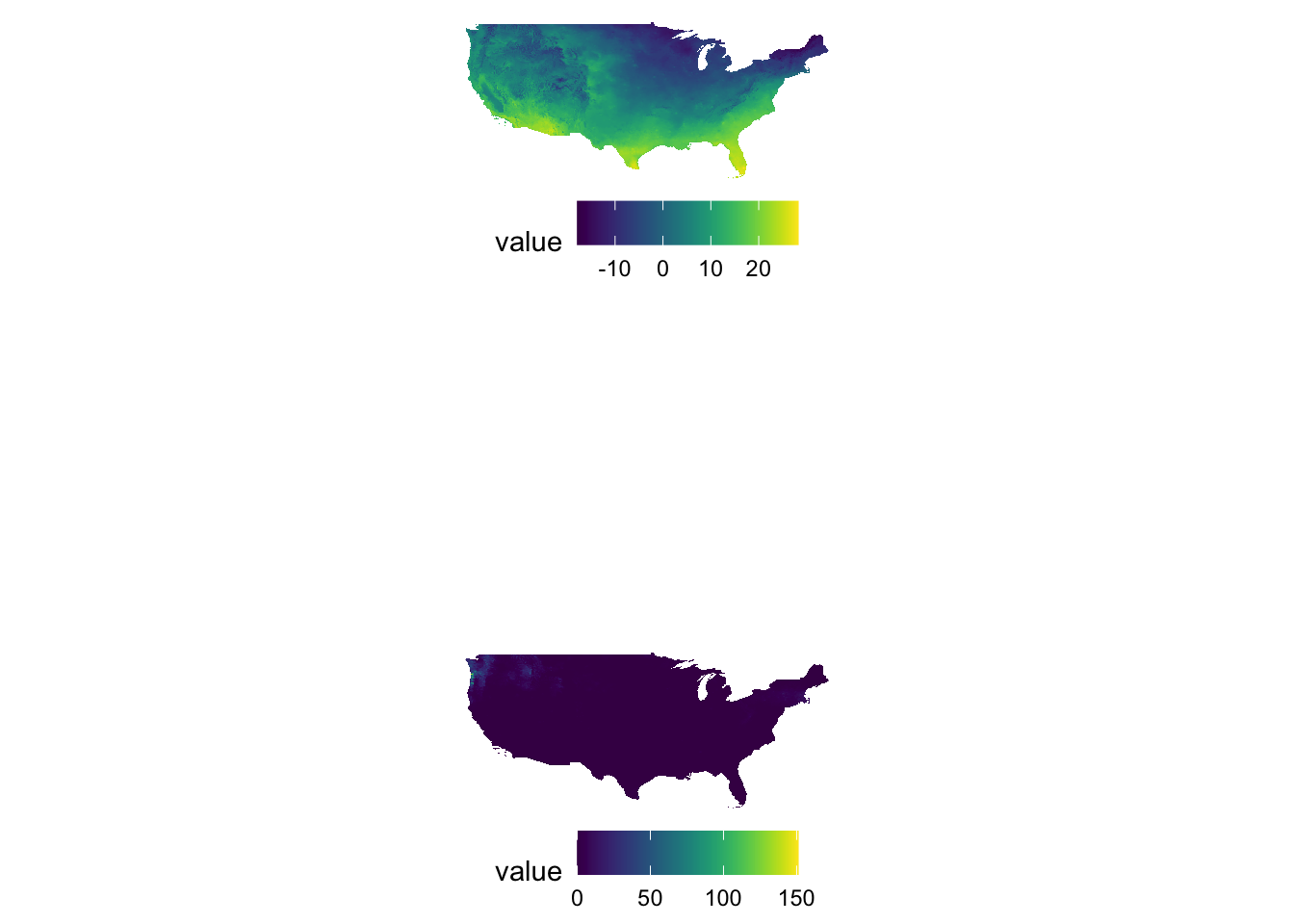
In plot.margin = unit(c(a, b, c, d), "cm"), here is which margin a, b, c, and d refers to.
- a: top
- b: right
- c: bottom
- d: left
8.4.2 A map in a map: inset
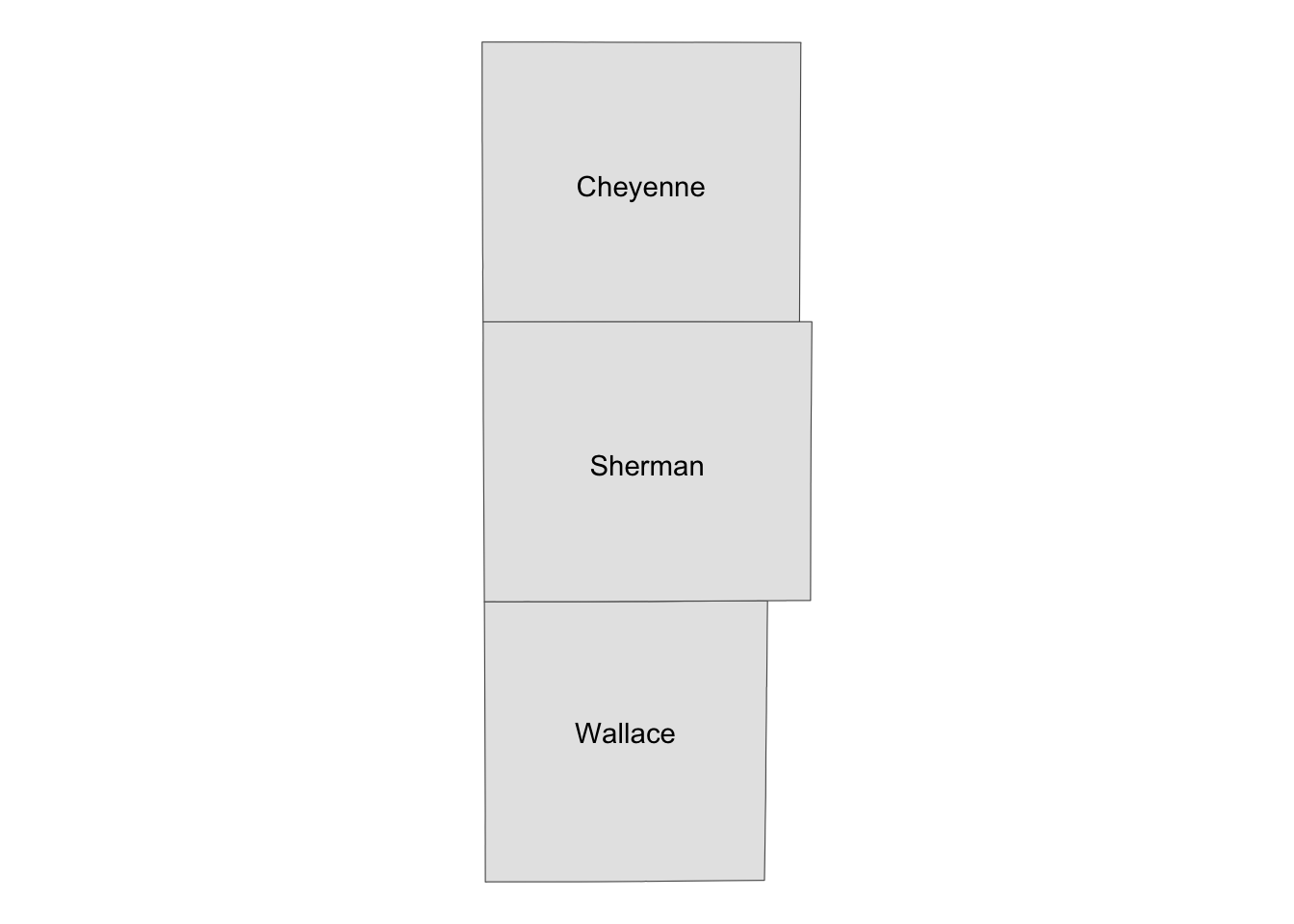
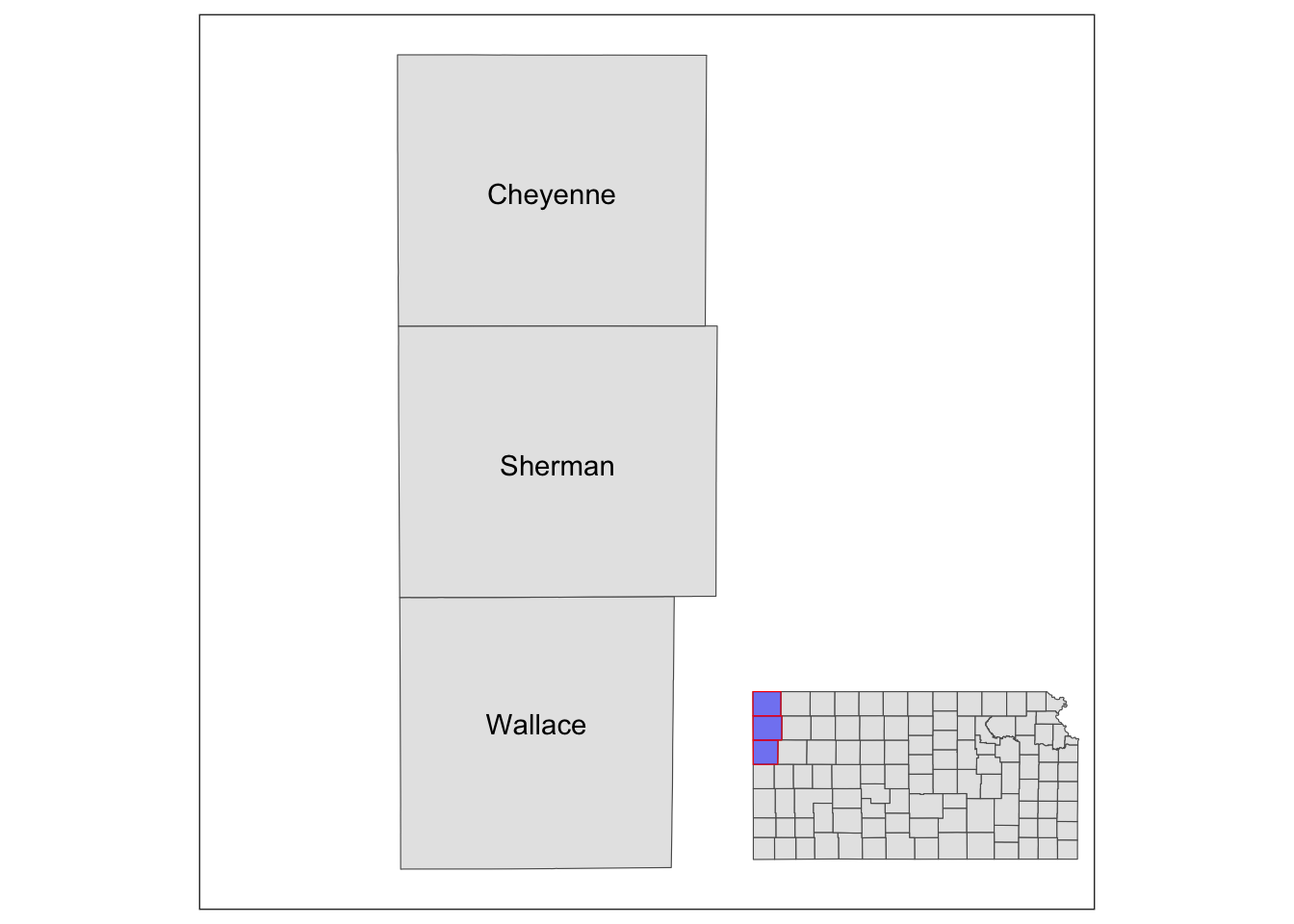
Sometimes, it is useful to present a map that covers a larger geographical range than the area of interest in the same map. This provides a better sense of the geographic extent and location of the area of interest relative to the larger geographic extent that the readers are more familiar with. For example, suppose your work is restricted to three counties in Kansas: Cheyenne, Sherman, and Wallace. Here is the map of three counties:
three_counties <- filter(KS_county, NAME %in% c("Cheyenne", "Sherman", "Wallace"))
(
g_three_counties <- ggplot() +
geom_sf(data = three_counties) +
geom_sf_text(data = three_counties, aes(label = NAME)) +
theme_void()
)
Well, for those who are not familiar with Kansas, it might be useful to show where in Kansas they are located on the same map (or even where Kansas is in the U.S.). This can be achieved using ggplotGrob() and annotation_custom(). The steps are the following:
- create a map of the area of interest and turn it into a
grobobject usingggplotGrob() - create a map of the region that includes the area of interest and turn it into a
grobobject usingggplotGrob() - combine the two using
annotation_custom()
#--- convert the ggplot into a grob ---#
grob_aoi <- ggplotGrob(g_three_counties)
#--- check the class ---#
class(grob_aoi)[1] "gtable" "gTree" "grob" "gDesc"
#--- create a map of Kansas ---#
g_region <- ggplot() +
geom_sf(data = KS_county) +
geom_sf(data = three_counties, fill = "blue", color = "red", alpha = 0.5) +
theme_void()
#--- convert to a grob ---#
grob_region <- ggplotGrob(g_region)Now that we have two maps, we can now put them on the same map using annotation_custom(). The first task is to initiate a ggplot with coord_equal() as follows:
(
g_inset <- ggplot() +
coord_equal(xlim = c(0, 1), ylim = c(0, 1), expand = FALSE)
)
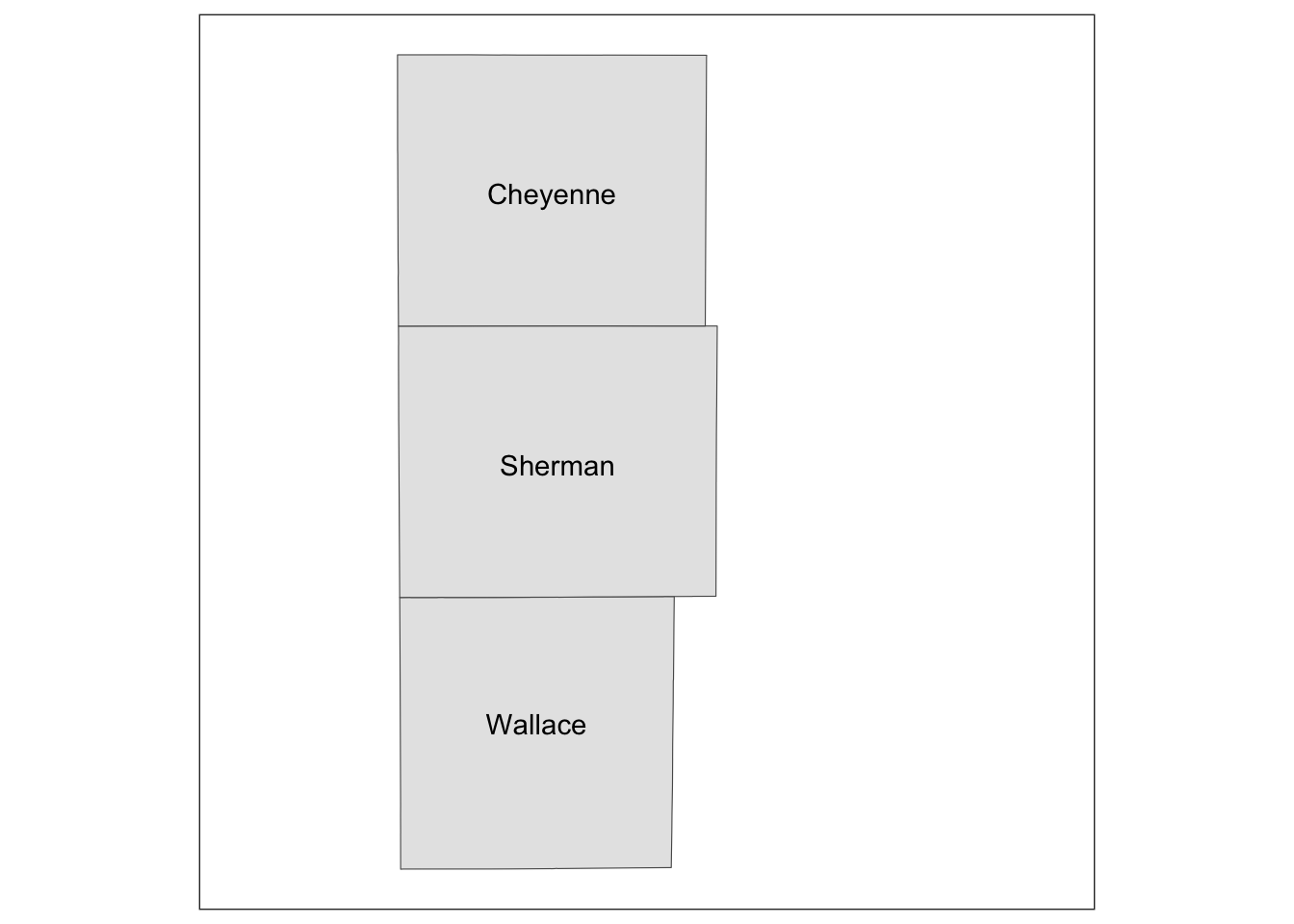
You now have a blank canvas to put the images on. Let’s add a layer to the canvas using annotation_custom() in which you provide the grob object (a map) and specify the range of the canvas the map occupies. Since the extent of x and y are set to [0, 1] above with coord_equal(xlim = c(0, 1), ylim = c(0, 1), expand = FALSE), the following code put the grob_aoi to cover the entire y range and up to 0.8 of x from 0.
g_inset +
annotation_custom(grob_aoi,
xmin = 0, xmax = 0.8, ymin = 0,
ymax = 1
)
Similarly, we can add grob_region using annotation_custom(). Let’s put it at the right lower corner of the map.
g_inset +
annotation_custom(grob_aoi,
xmin = 0, xmax = 0.8, ymin = 0,
ymax = 1
) +
annotation_custom(grob_region,
xmin = 0.6, xmax = 1, ymin = 0,
ymax = 0.3
)
Note that the resulting map still has the default theme because it does not inherit the theme of maps added by annotation_custom(). So, you can add theme_void() to the map to make the border disappear.
g_inset +
annotation_custom(grob_aoi,
xmin = 0, xmax = 0.8, ymin = 0,
ymax = 1
) +
annotation_custom(grob_region,
xmin = 0.6, xmax = 1, ymin = 0,
ymax = 0.3
) +
theme_void()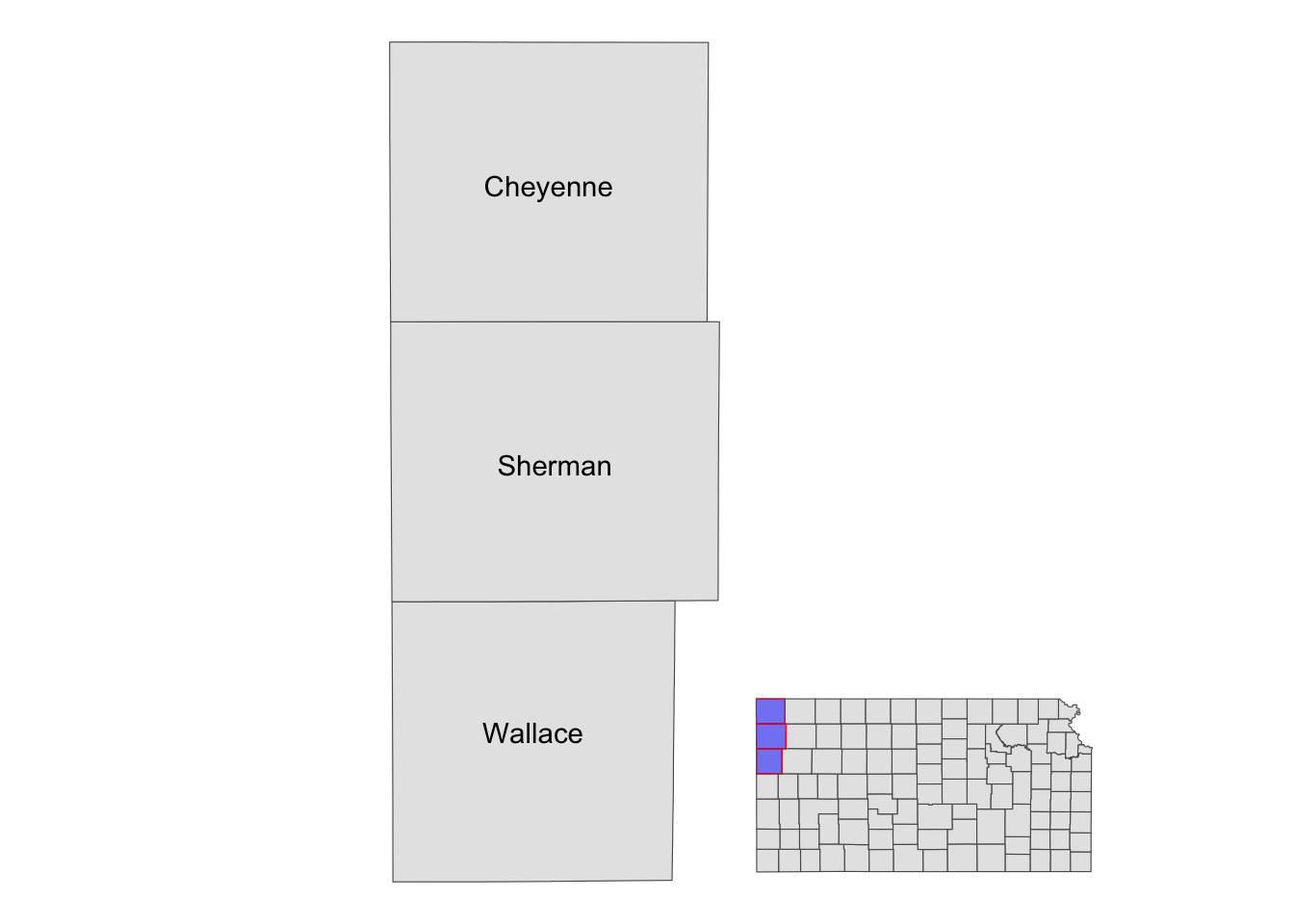
8.5 Fine-tuning maps for publication
This section presents a number of small tips to beautify your maps so that they can be publishable in professional reports. Often times, academic journals have their own particular sets of rules about figures, and reviewers or your boss might have their own views of what maps should look like. Whatever the requirements or requests, you need to accommodate their requests and modify the maps accordingly. It is worth mentioning that there is really nothing specific to creating maps here. Techniques presented here are applicable to any kind of figure. It is just that we limit ourselves to specific components of a figure that you are likely to want to modify from the default when creating maps. Specifically, pre-made ggplot2 themes and how to modify legends and facet strips are discussed.
(
gw_by_county <- st_join(KS_county, gw_KS_sf) %>%
data.table() %>%
.[, .(af_used = sum(af_used, na.rm = TRUE)), by = .(COUNTYFP, year)] %>%
left_join(KS_county, ., by = c("COUNTYFP")) %>%
filter(!is.na(year))
)Simple feature collection with 184 features and 14 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -102.0517 ymin: 36.99302 xmax: -94.58841 ymax: 40.00316
Geodetic CRS: NAD83
First 10 features:
STATEFP COUNTYFP COUNTYNS AFFGEOID GEOID NAME NAMELSAD
1 20 175 00485050 0500000US20175 20175 Seward Seward County
2 20 175 00485050 0500000US20175 20175 Seward Seward County
3 20 027 00484983 0500000US20027 20027 Clay Clay County
4 20 027 00484983 0500000US20027 20027 Clay Clay County
5 20 171 00485048 0500000US20171 20171 Scott Scott County
6 20 171 00485048 0500000US20171 20171 Scott Scott County
7 20 047 00484993 0500000US20047 20047 Edwards Edwards County
8 20 047 00484993 0500000US20047 20047 Edwards Edwards County
9 20 147 00485037 0500000US20147 20147 Phillips Phillips County
10 20 147 00485037 0500000US20147 20147 Phillips Phillips County
STUSPS STATE_NAME LSAD ALAND AWATER year af_used
1 KS Kansas 06 1656693304 1961444 2010 163703.719
2 KS Kansas 06 1656693304 1961444 2011 218863.983
3 KS Kansas 06 1671314413 26701337 2010 5929.143
4 KS Kansas 06 1671314413 26701337 2011 8358.903
5 KS Kansas 06 1858536838 306079 2010 42542.602
6 KS Kansas 06 1858536838 306079 2011 51620.454
7 KS Kansas 06 1610699245 206413 2010 109726.389
8 KS Kansas 06 1610699245 206413 2011 141487.791
9 KS Kansas 06 2294395636 22493383 2010 4657.746
10 KS Kansas 06 2294395636 22493383 2011 5076.198
geometry
1 MULTIPOLYGON (((-101.0681 3...
2 MULTIPOLYGON (((-101.0681 3...
3 MULTIPOLYGON (((-97.3707 39...
4 MULTIPOLYGON (((-97.3707 39...
5 MULTIPOLYGON (((-101.1284 3...
6 MULTIPOLYGON (((-101.1284 3...
7 MULTIPOLYGON (((-99.56988 3...
8 MULTIPOLYGON (((-99.56988 3...
9 MULTIPOLYGON (((-99.62821 3...
10 MULTIPOLYGON (((-99.62821 3...
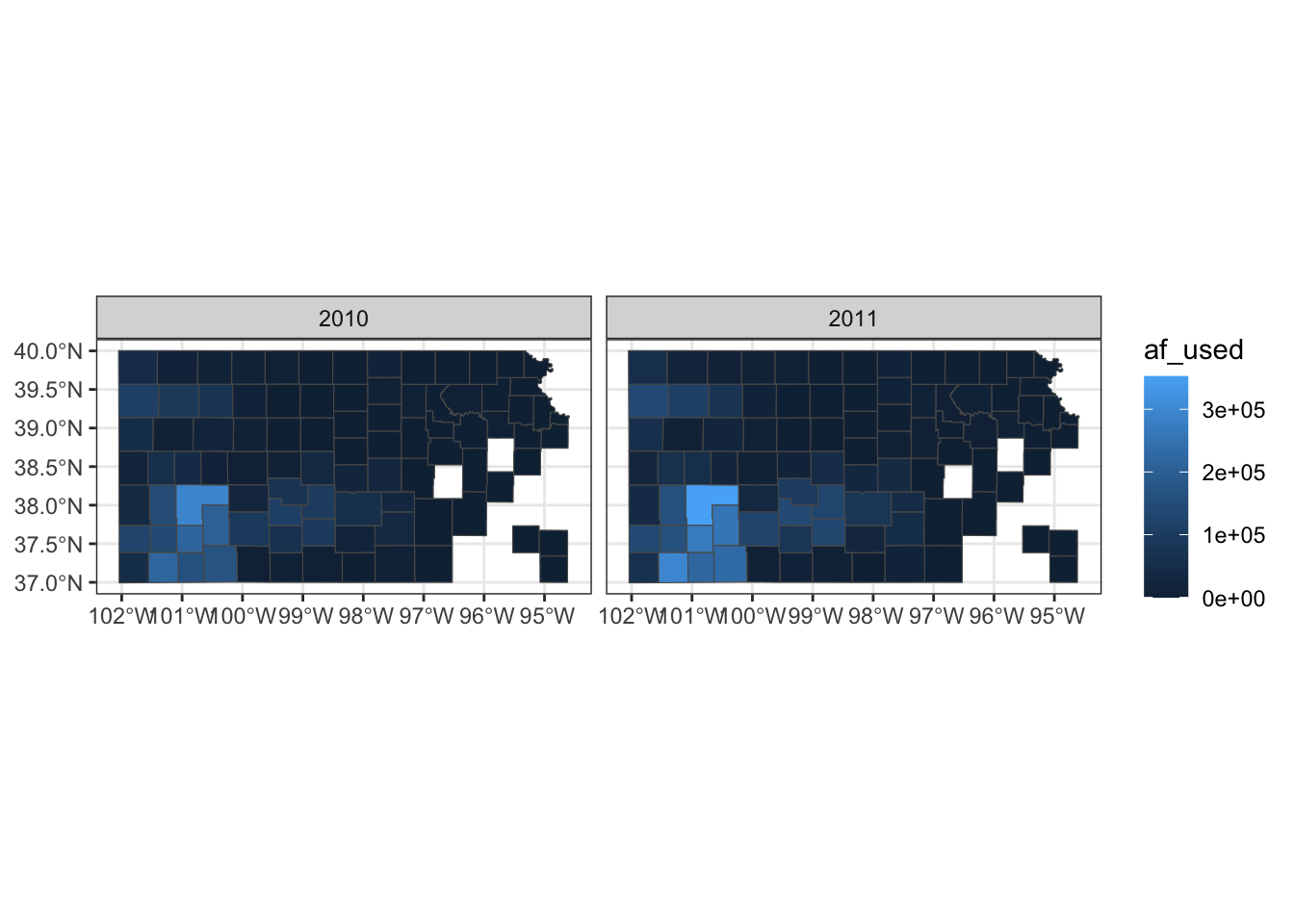
(
g_base <- ggplot() +
geom_sf(data = gw_by_county, aes(fill = af_used)) +
facet_wrap(. ~ year)
)
8.5.1 Setting the theme
Right now, the map shows geographic coordinates, gray background, and grid lines. They are not very aesthetically appealing. Adding the pre-defined theme by theme_*() can alter the theme of a map very quickly. One of the themes suitable for maps is theme_void().
g_base + theme_void()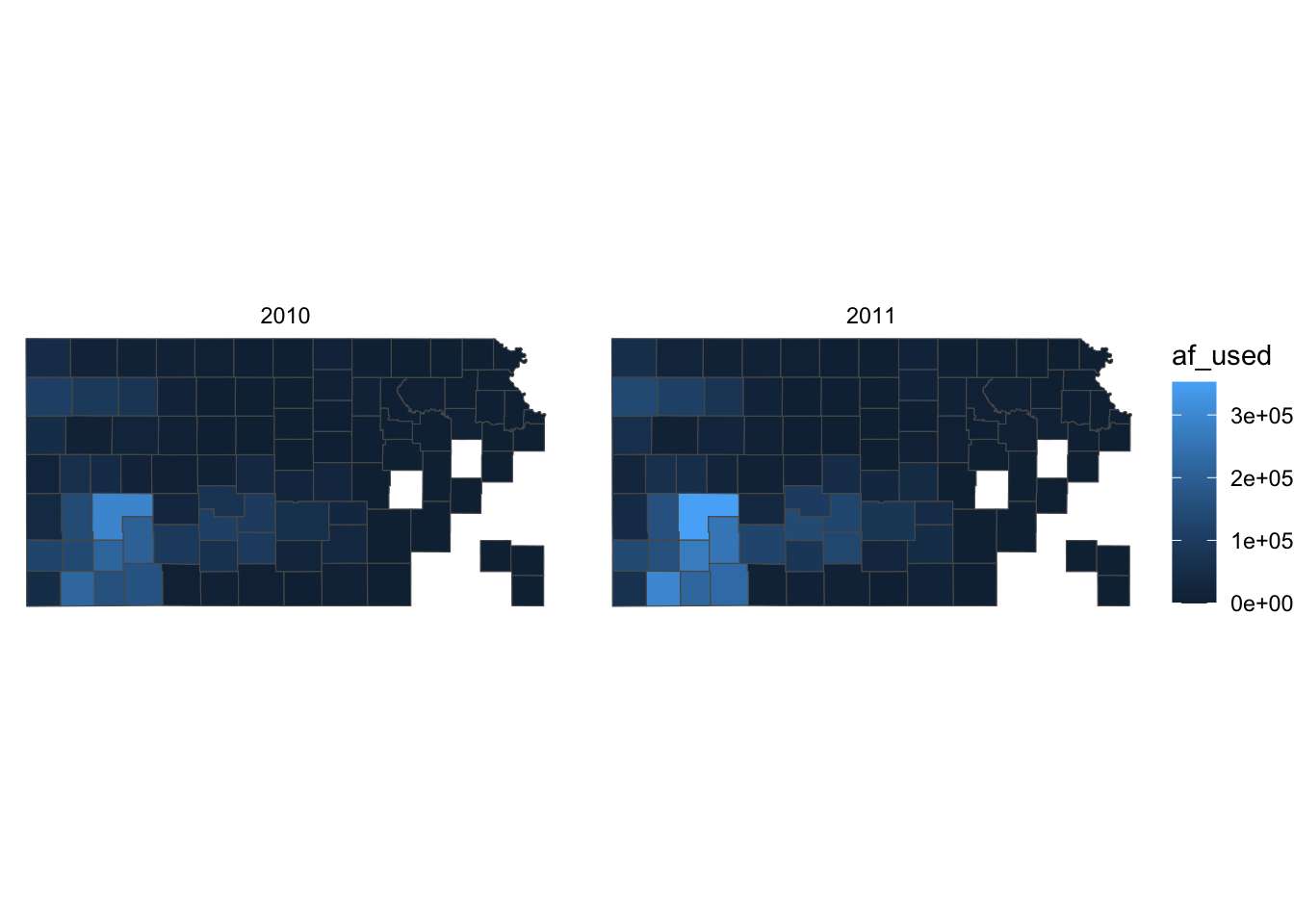
As you can see, all the axes information (axis.title, axis.ticks, axis.text,) and panel information (panel.background, panel.border, panel.grid) are gone among other parts of the figure. You can confirm this by evaluating theme_void().
List of 136
$ line : list()
..- attr(*, "class")= chr [1:2] "element_blank" "element"
$ rect : list()
..- attr(*, "class")= chr [1:2] "element_blank" "element"
$ text :List of 11
..$ family : chr ""
..$ face : chr "plain"
..$ colour : chr "black"
..$ size : num 11
..$ hjust : num 0.5
..$ vjust : num 0.5
..$ angle : num 0
..$ lineheight : num 0.9
..$ margin : 'margin' num [1:4] 0points 0points 0points 0points
.. ..- attr(*, "unit")= int 8
..$ debug : logi FALSE
..$ inherit.blank: logi TRUE
..- attr(*, "class")= chr [1:2] "element_text" "element"
$ title : NULL
$ aspect.ratio : NULL
$ axis.title : list()
..- attr(*, "class")= chr [1:2] "element_blank" "element"
$ axis.title.x : NULL
$ axis.title.x.top : NULL
$ axis.title.x.bottom : NULL
$ axis.title.y : NULL
$ axis.title.y.left : NULL
$ axis.title.y.right : NULL
$ axis.text : list()
..- attr(*, "class")= chr [1:2] "element_blank" "element"
$ axis.text.x : NULL
$ axis.text.x.top : NULL
$ axis.text.x.bottom : NULL
$ axis.text.y : NULL
$ axis.text.y.left : NULL
$ axis.text.y.right : NULL
$ axis.text.theta : NULL
$ axis.text.r : NULL
$ axis.ticks : NULL
$ axis.ticks.x : NULL
$ axis.ticks.x.top : NULL
$ axis.ticks.x.bottom : NULL
$ axis.ticks.y : NULL
$ axis.ticks.y.left : NULL
$ axis.ticks.y.right : NULL
$ axis.ticks.theta : NULL
$ axis.ticks.r : NULL
$ axis.minor.ticks.x.top : NULL
$ axis.minor.ticks.x.bottom : NULL
$ axis.minor.ticks.y.left : NULL
$ axis.minor.ticks.y.right : NULL
$ axis.minor.ticks.theta : NULL
$ axis.minor.ticks.r : NULL
$ axis.ticks.length : 'simpleUnit' num 0points
..- attr(*, "unit")= int 8
$ axis.ticks.length.x : NULL
$ axis.ticks.length.x.top : NULL
$ axis.ticks.length.x.bottom : NULL
$ axis.ticks.length.y : NULL
$ axis.ticks.length.y.left : NULL
$ axis.ticks.length.y.right : NULL
$ axis.ticks.length.theta : NULL
$ axis.ticks.length.r : NULL
$ axis.minor.ticks.length : 'simpleUnit' num 0points
..- attr(*, "unit")= int 8
$ axis.minor.ticks.length.x : NULL
$ axis.minor.ticks.length.x.top : NULL
$ axis.minor.ticks.length.x.bottom: NULL
$ axis.minor.ticks.length.y : NULL
$ axis.minor.ticks.length.y.left : NULL
$ axis.minor.ticks.length.y.right : NULL
$ axis.minor.ticks.length.theta : NULL
$ axis.minor.ticks.length.r : NULL
$ axis.line : NULL
$ axis.line.x : NULL
$ axis.line.x.top : NULL
$ axis.line.x.bottom : NULL
$ axis.line.y : NULL
$ axis.line.y.left : NULL
$ axis.line.y.right : NULL
$ axis.line.theta : NULL
$ axis.line.r : NULL
$ legend.background : NULL
$ legend.margin : NULL
$ legend.spacing : NULL
$ legend.spacing.x : NULL
$ legend.spacing.y : NULL
$ legend.key : NULL
$ legend.key.size : 'simpleUnit' num 1.2lines
..- attr(*, "unit")= int 3
$ legend.key.height : NULL
$ legend.key.width : NULL
$ legend.key.spacing : 'simpleUnit' num 5.5points
..- attr(*, "unit")= int 8
$ legend.key.spacing.x : NULL
$ legend.key.spacing.y : NULL
$ legend.frame : NULL
$ legend.ticks : NULL
$ legend.ticks.length : 'rel' num 0.2
$ legend.axis.line : NULL
$ legend.text :List of 11
..$ family : NULL
..$ face : NULL
..$ colour : NULL
..$ size : 'rel' num 0.8
..$ hjust : NULL
..$ vjust : NULL
..$ angle : NULL
..$ lineheight : NULL
..$ margin : NULL
..$ debug : NULL
..$ inherit.blank: logi TRUE
..- attr(*, "class")= chr [1:2] "element_text" "element"
$ legend.text.position : NULL
$ legend.title :List of 11
..$ family : NULL
..$ face : NULL
..$ colour : NULL
..$ size : NULL
..$ hjust : num 0
..$ vjust : NULL
..$ angle : NULL
..$ lineheight : NULL
..$ margin : NULL
..$ debug : NULL
..$ inherit.blank: logi TRUE
..- attr(*, "class")= chr [1:2] "element_text" "element"
$ legend.title.position : NULL
$ legend.position : chr "right"
$ legend.position.inside : NULL
$ legend.direction : NULL
$ legend.byrow : NULL
$ legend.justification : NULL
$ legend.justification.top : NULL
$ legend.justification.bottom : NULL
$ legend.justification.left : NULL
$ legend.justification.right : NULL
$ legend.justification.inside : NULL
$ legend.location : NULL
$ legend.box : NULL
$ legend.box.just : NULL
$ legend.box.margin : NULL
$ legend.box.background : NULL
$ legend.box.spacing : NULL
[list output truncated]
- attr(*, "class")= chr [1:2] "theme" "gg"
- attr(*, "complete")= logi TRUE
- attr(*, "validate")= logi TRUEApplying the theme to a map obviates the need to suppress parts of a figure individually. You can suppress parts of the figure individually using theme(). For example, the following code gets rid of axis.text.
g_base + theme(axis.text = element_blank())
So, if theme_void() is overdoing things, you can build your own theme specifically for maps. For example, this is the theme I used for maps in Chapter 1.
theme_for_map <-
theme(
axis.ticks = element_blank(),
axis.text = element_blank(),
axis.line = element_blank(),
panel.border = element_blank(),
panel.grid = element_line(color = "transparent"),
panel.background = element_blank(),
plot.background = element_rect(fill = "transparent", color = "transparent")
)Applying the theme to the map:
g_base + theme_for_map
This is very similar to theme_void() except that strip.background is not lost.
You can use theme_void() as a starting point and override components of it like this.
#--- bring back a color to strip.background ---#
theme_for_map_2 <- theme_void() + theme(strip.background = element_rect(fill = "gray"))
#--- apply the new theme ---#
g_base + theme_for_map_2
theme_bw() is also a good theme for maps.
ggplot() +
geom_sf(data = gw_by_county, aes(fill = af_used)) +
facet_grid(year ~ .) +
theme_bw()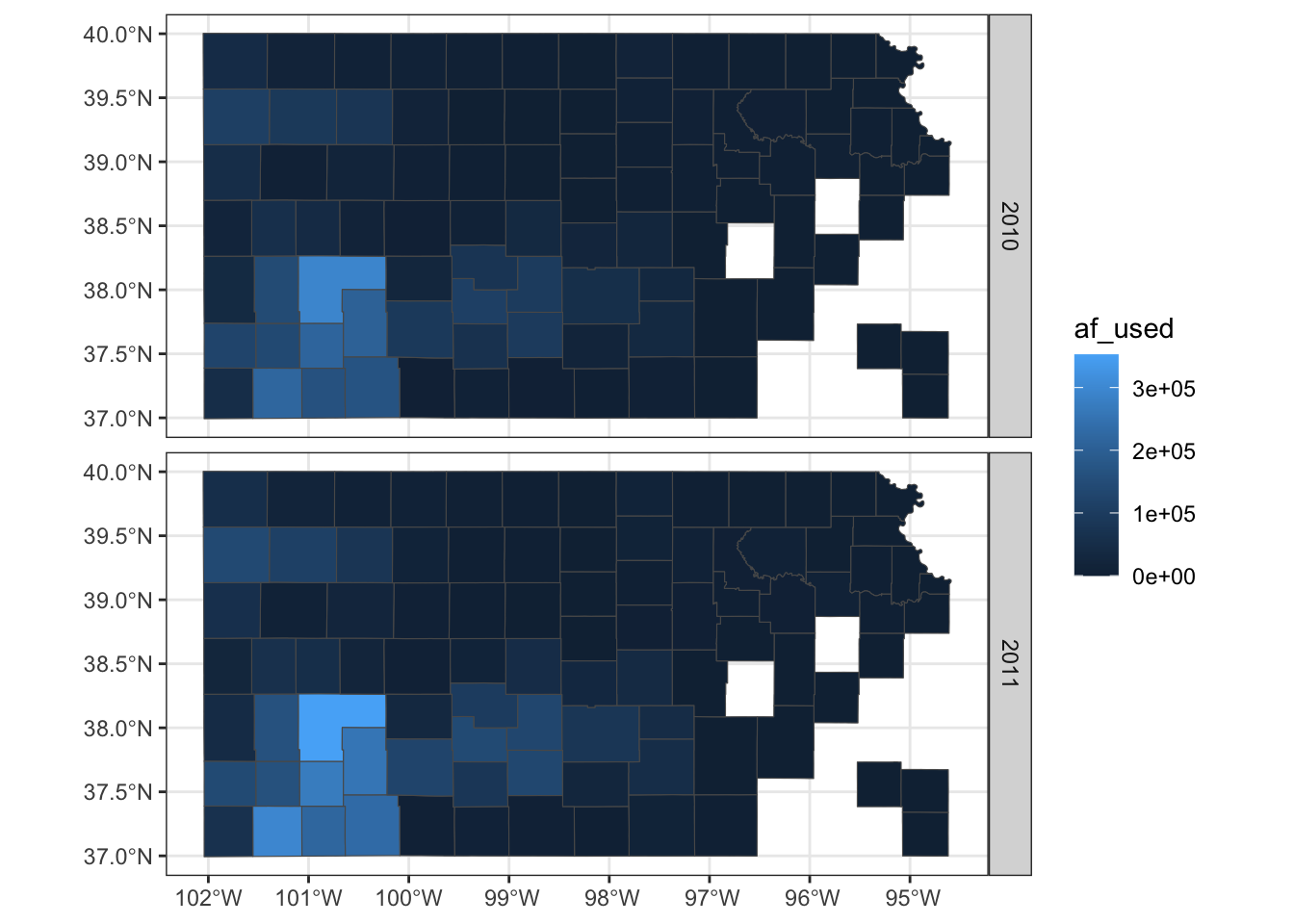
If you do not like the gray grid lines, you can remove them like this.
ggplot() +
geom_sf(data = gw_by_county, aes(fill = af_used)) +
facet_grid(year ~ .) +
theme_bw() +
theme(
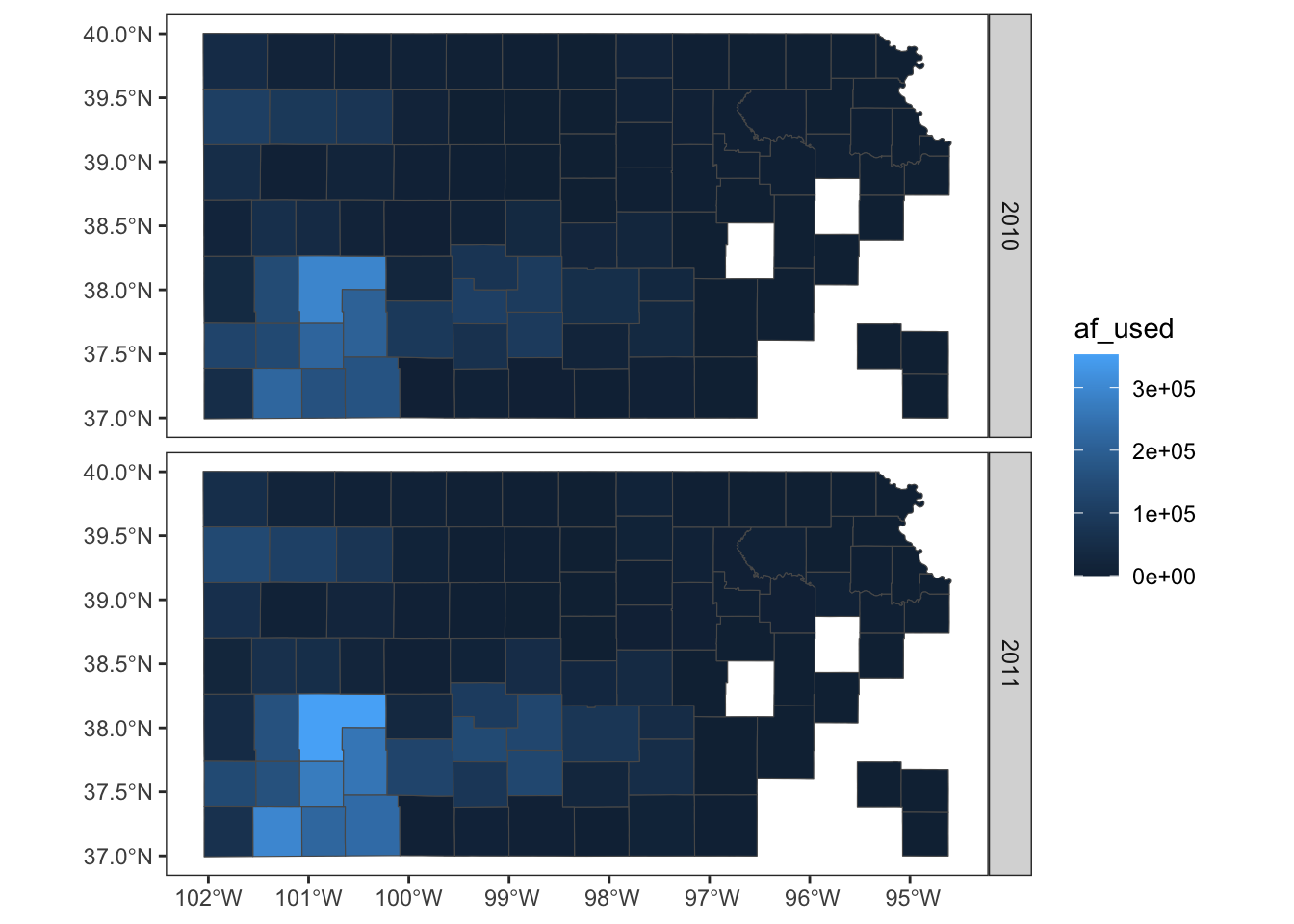
panel.grid = element_line(color = "transparent")
)
Not all themes are suitable for maps. For example, theme_classic() is not a very good option as you can see below:
g_base + theme_classic()
If you are not satisfied with theme_void() and not willing to make up your own theme, then you may want to take a look at other pre-made themes that are available from ggplot2 (see here) and ggthemes (see here). Note that some themes are more invasive than theme_void(), altering the default color scale.
8.5.2 Legend
Legends can be modified using legend.*() options for theme() and guide_*(). It is impossible to discuss every single one of all the options for these functions. So, this section focuses on the most common and useful (that I consider) modifications you can make to legends.
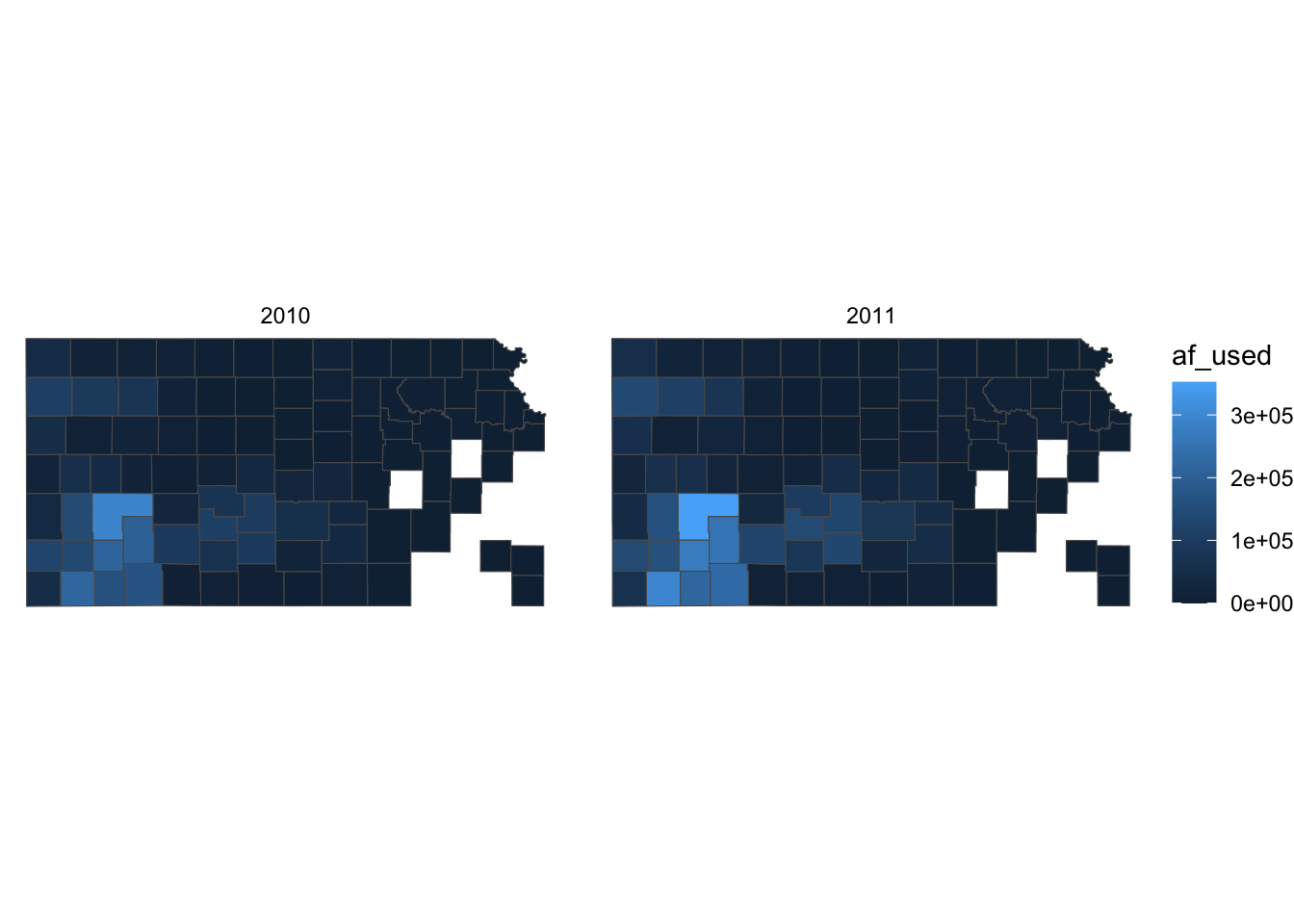
A legend consists of three elements: legend title, legend key (e.g., color bar), and legend label (or legend text). For example, in the figure below, af_used is the legend title, the color bar is the legend key, and the numbers below the color bar are legend labels. Knowing the name of these elements helps because the name of the options contains the name of the specific part of the legend.
(
g_legend <- ggplot() +
geom_sf(data = gw_by_county, aes(fill = af_used)) +
facet_wrap(. ~ year) +
theme_void()
)
Let’s first change the color scale to Viridis using scale_fill_viridis_c() (see section 8.3 for picking a color scale).
g_legend +
scale_fill_viridis_c()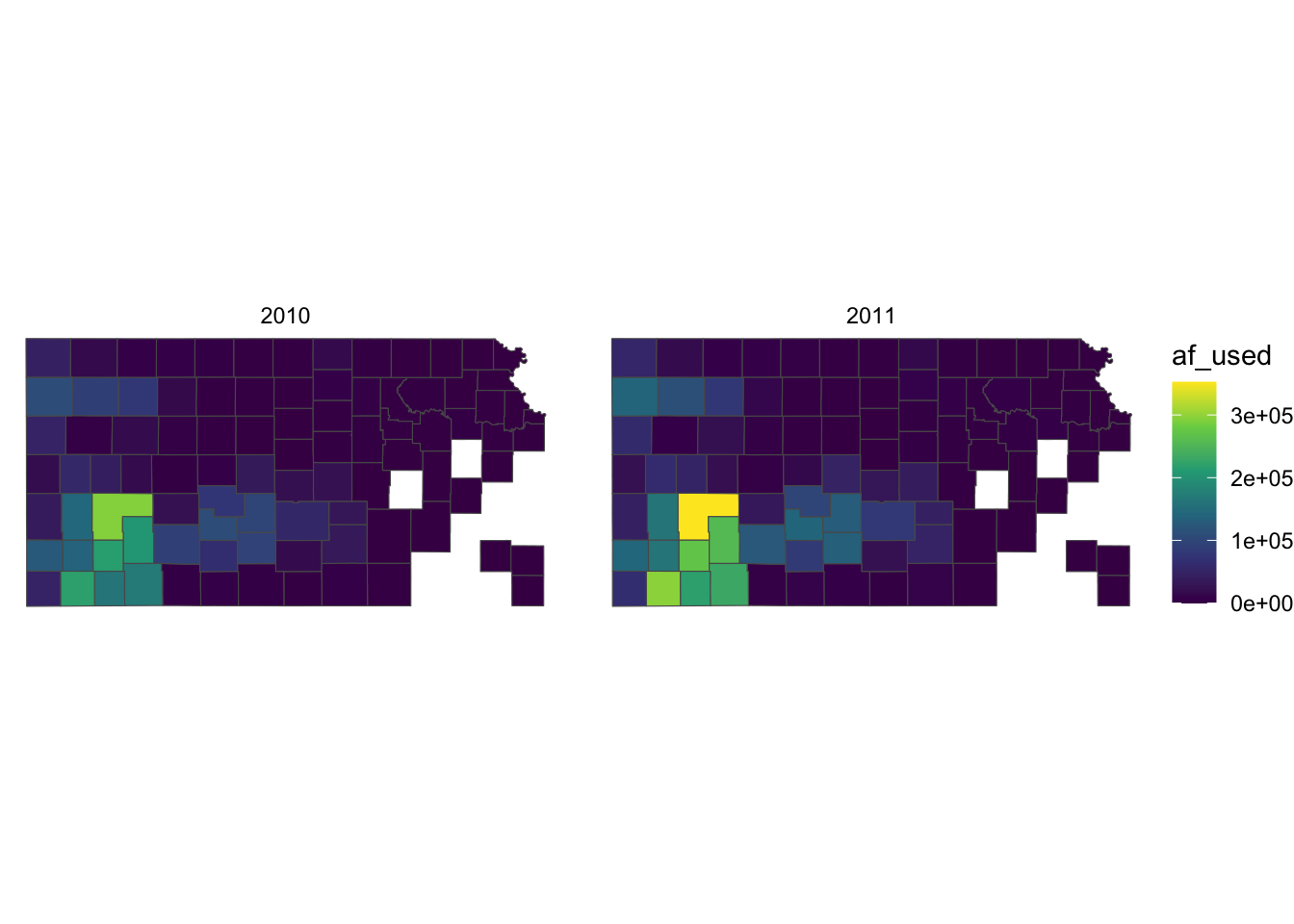
Right now, the legend title is af_used, which does not tell the readers what it means. In general, you can change the title of a legend by using the name option inside the scale function for the legend (here, scale_fill_viridis_c()). So, this one works:
g_legend +
scale_fill_viridis_c(name = "Groundwater pumping (acre-feet)")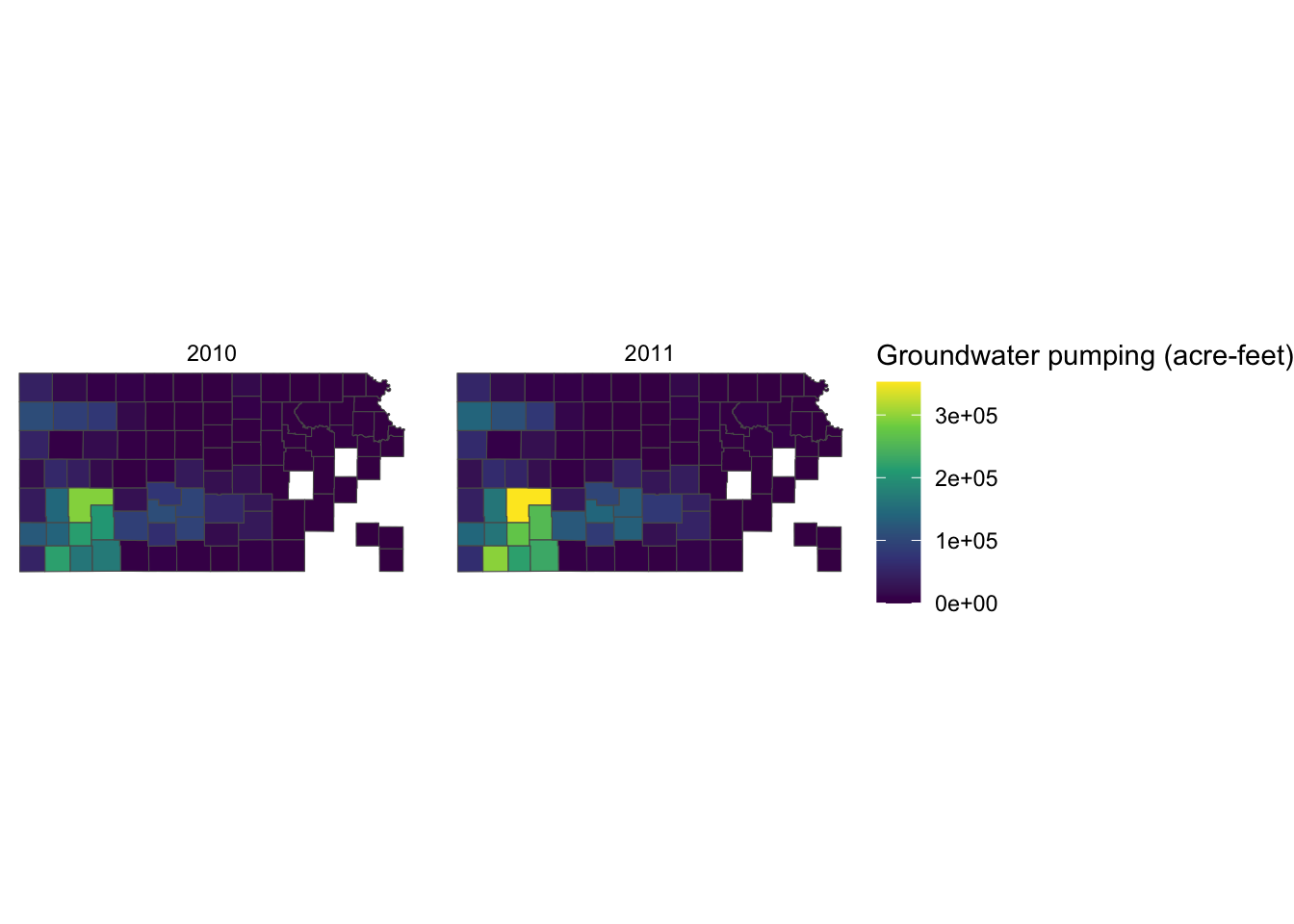
Alternatively, you can use labs() function.93. Since the legend is for the fill aesthetic attribute, you should add fill = "legend title" as follows:
g_legend +
scale_fill_viridis_c() +
labs(fill = "Groundwater pumping (acre-feet)")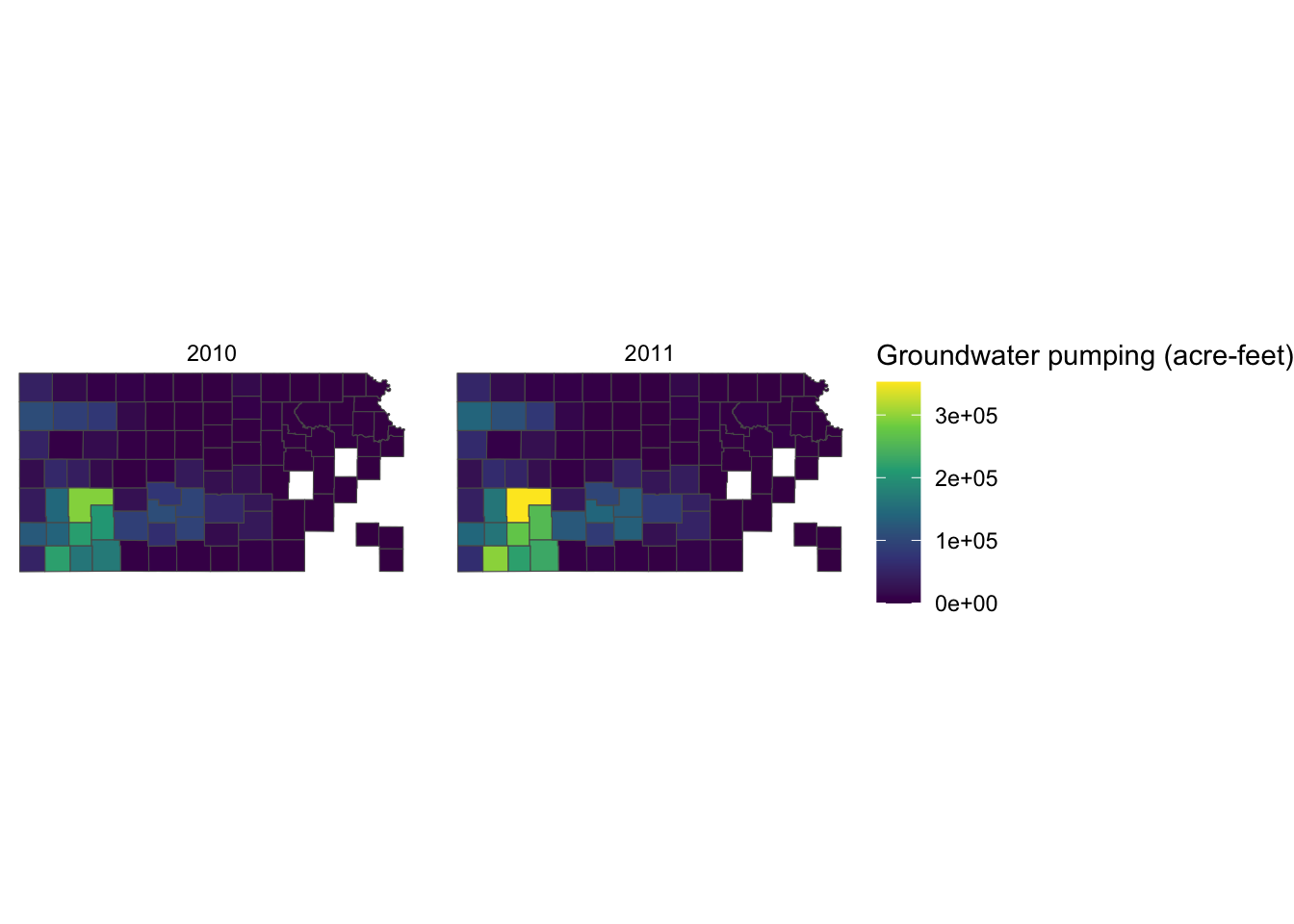
Since the legend title is long, the legend is taking up about the half of the space of the entire figure. So, let’s put the legend below the maps (bottom of the figure) by adding theme(legend.position = "bottom").
g_legend +
scale_fill_viridis_c() +
labs(fill = "Groundwater pumping (acre-feet)") +
theme(legend.position = "bottom")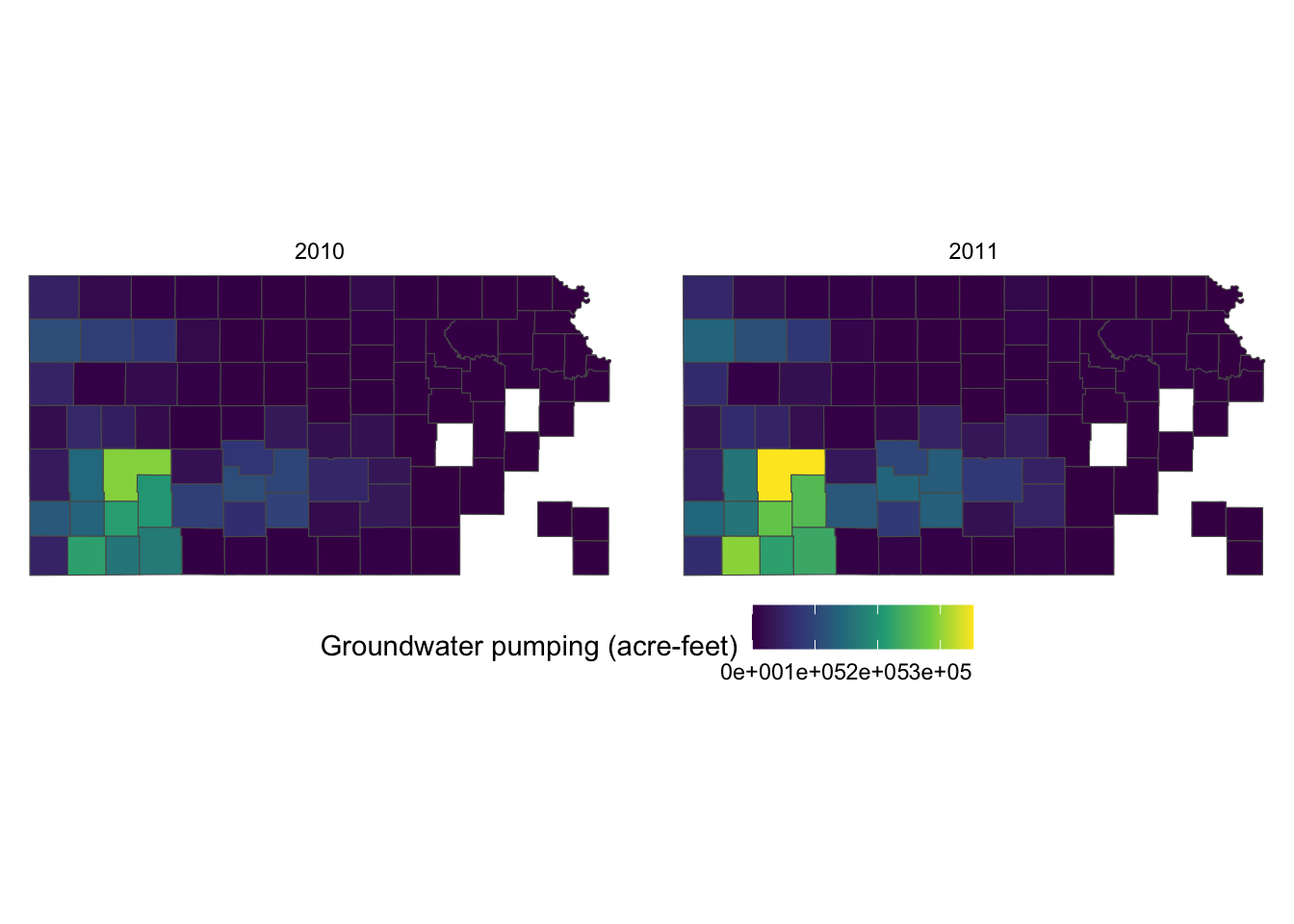
It would be aesthetically better to have the legend title on top of the color bar. This can be done by using the guides() function. Since we would like to alter the aesthetics of the legend for fill involving a color bar, we use fill = guide_colorbar(). To place the legend title on top, you add title.position="top" inside the guide_colorbar() function as follows:
g_legend +
scale_fill_viridis_c() +
labs(fill = "Groundwater pumping (acre-feet)") +
theme(legend.position = "bottom") +
guides(fill = guide_colorbar(title.position = "top"))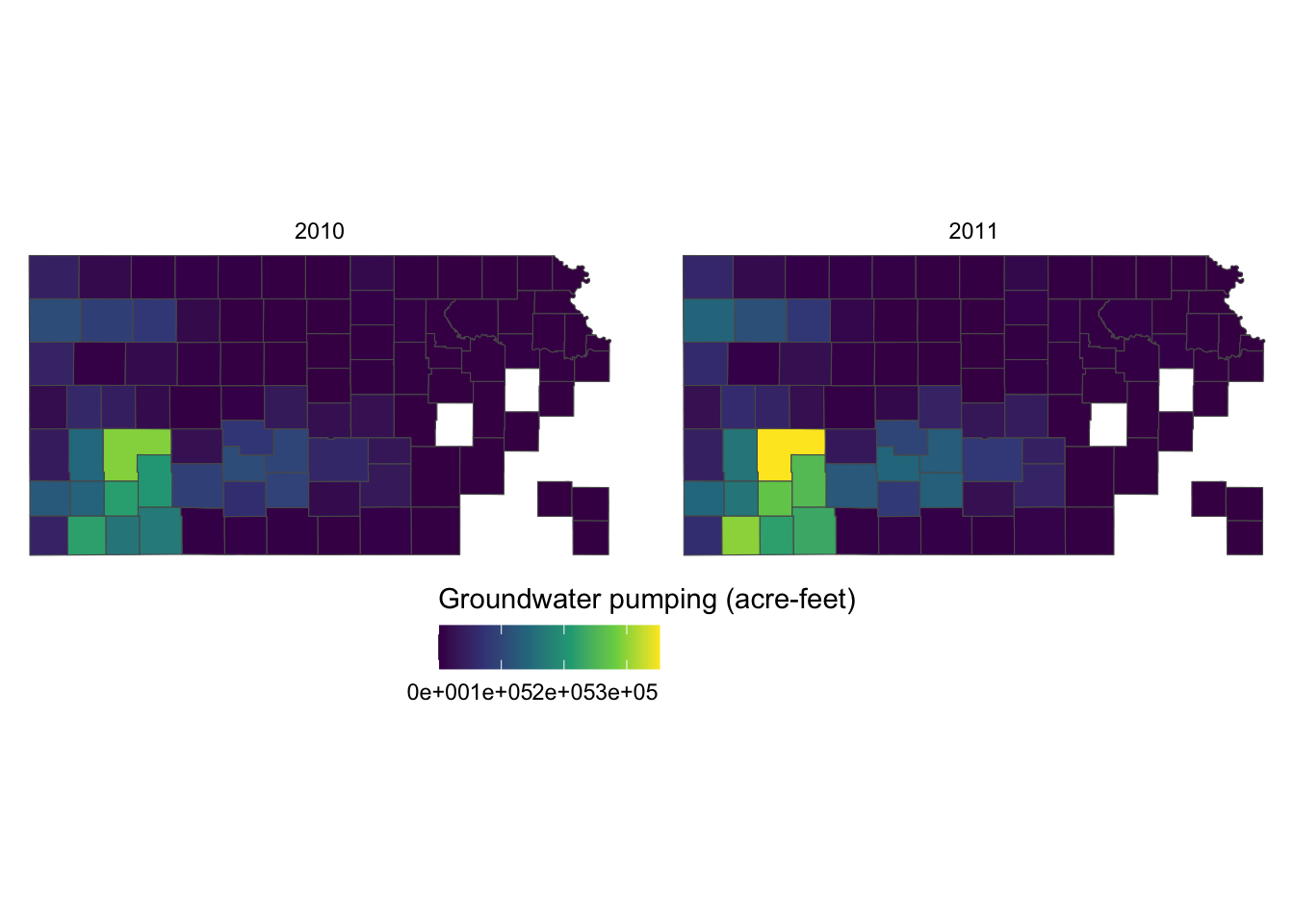
This looks better. But, the legend labels are too close to each other so it is hard to read them because the color bar is too short. Let’s elongate the color bar so that we have enough space between legend labels using legend.key.width = option for theme(). Let’s also make the legend thinner using legend.key.height = option.
g_legend +
scale_fill_viridis_c() +
labs(fill = "Groundwater pumping (acre-feet)") +
theme(
legend.position = "bottom",
#--- NEW LINES HERE!! ---#
legend.key.height = unit(0.5, "cm"),
legend.key.width = unit(2, "cm")
) +
guides(fill = guide_colorbar(title.position = "top"))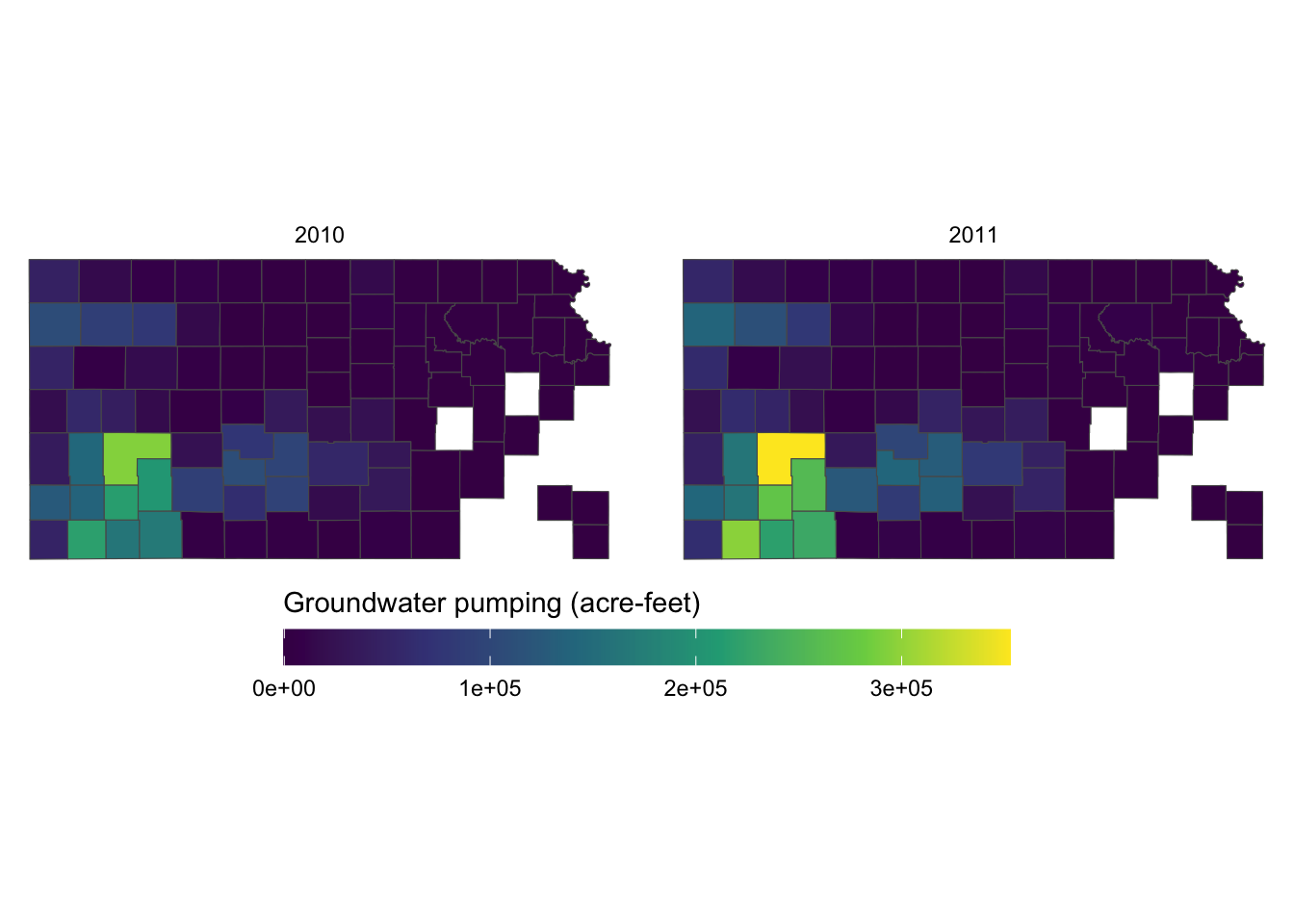
If the journal you are submitting an article to is requesting a specific font family for the texts in the figure, you can use legend.text = element_text() and legend.title = element_text() inside theme() for the legend labels and legend title, respectively. The following code uses the font family of “Times” and font size of 12 for both the labels and the title.
g_legend +
scale_fill_viridis_c() +
labs(fill = "Groundwater pumping (acre-feet)") +
theme(
legend.position = "bottom",
legend.key.height = unit(0.5, "cm"),
legend.key.width = unit(2, "cm"),
legend.text = element_text(size = 12, family = "Times"),
legend.title = element_text(size = 12, family = "Times")
#--- NEW LINES HERE!! ---#
) +
guides(fill = guide_colorbar(title.position = "top"))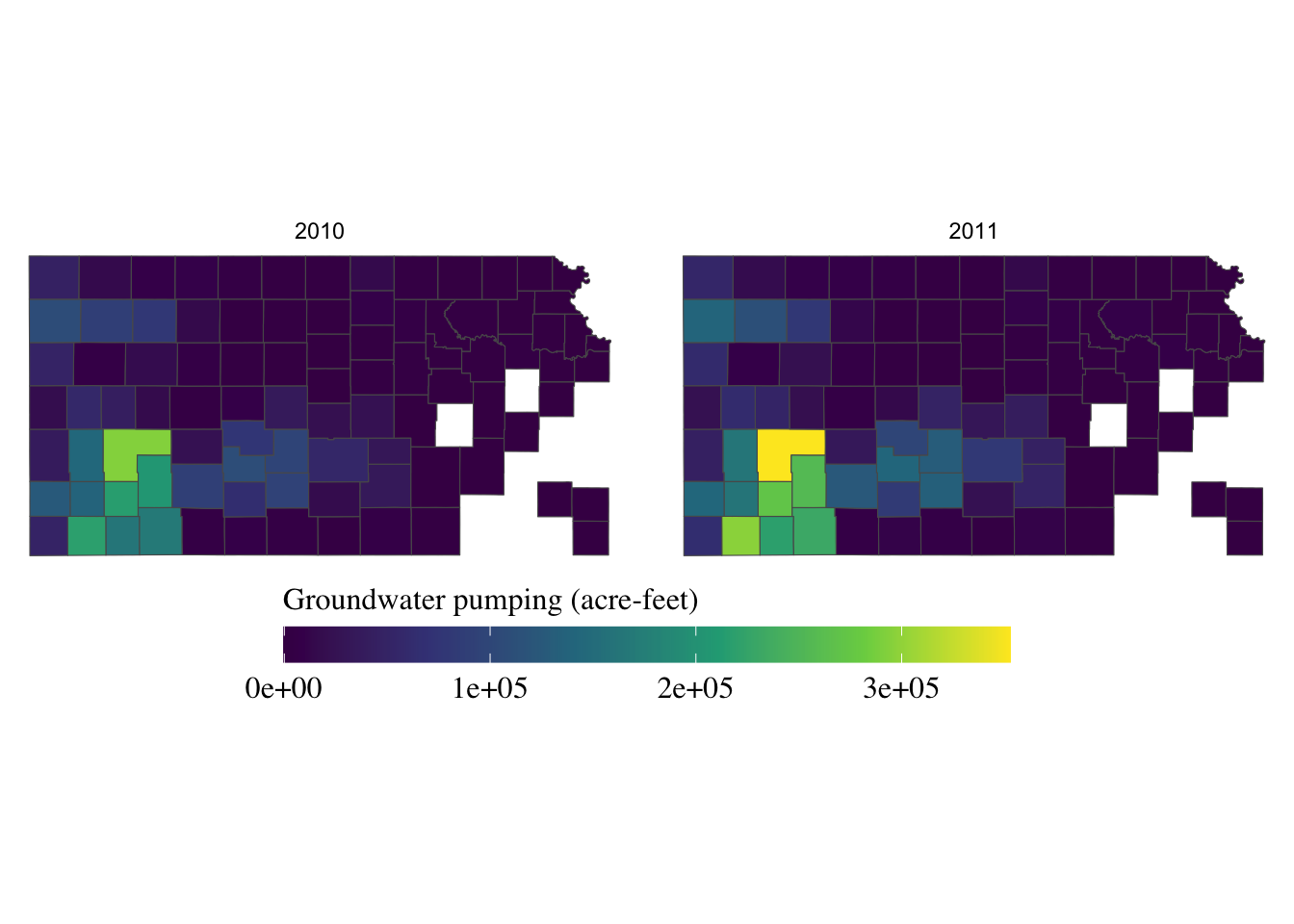
For the other options to control the legend with a color bar, see here.
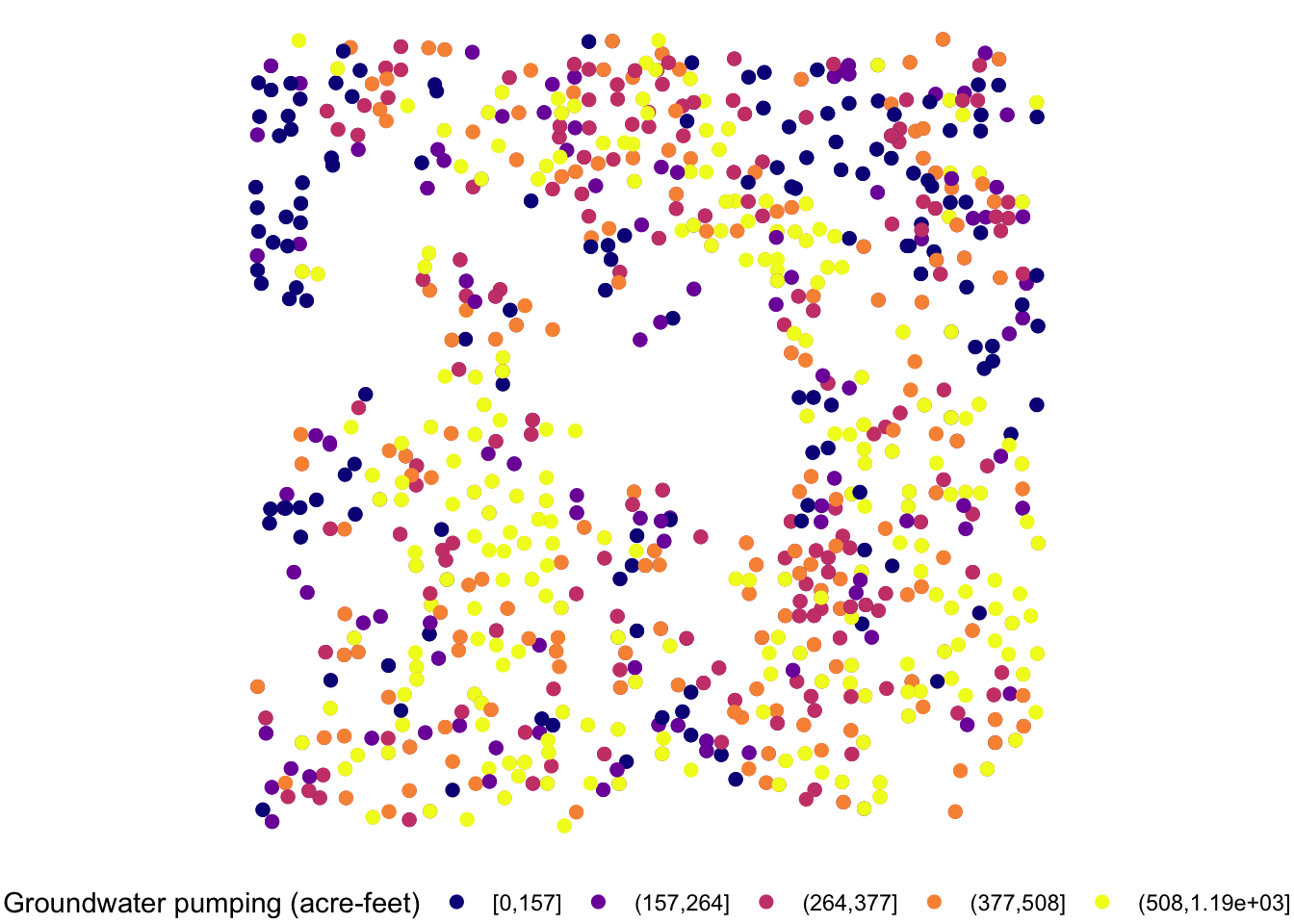
When the legend is made for discrete values, you can use guide_legend(). Let’s use the following map as a starting point.
#--- convert af_used to a discrete variable ---#
gw_Stevens <- mutate(gw_Stevens, af_used_cat = cut_number(af_used, n = 5))
(
g_legend_2 <-
ggplot(data = gw_Stevens) +
geom_sf(aes(color = af_used_cat), size = 2) +
scale_color_viridis(discrete = TRUE, option = "C") +
labs(color = "Groundwater pumping (acre-feet)") +
theme_void() +
theme(legend.position = "bottom")
)
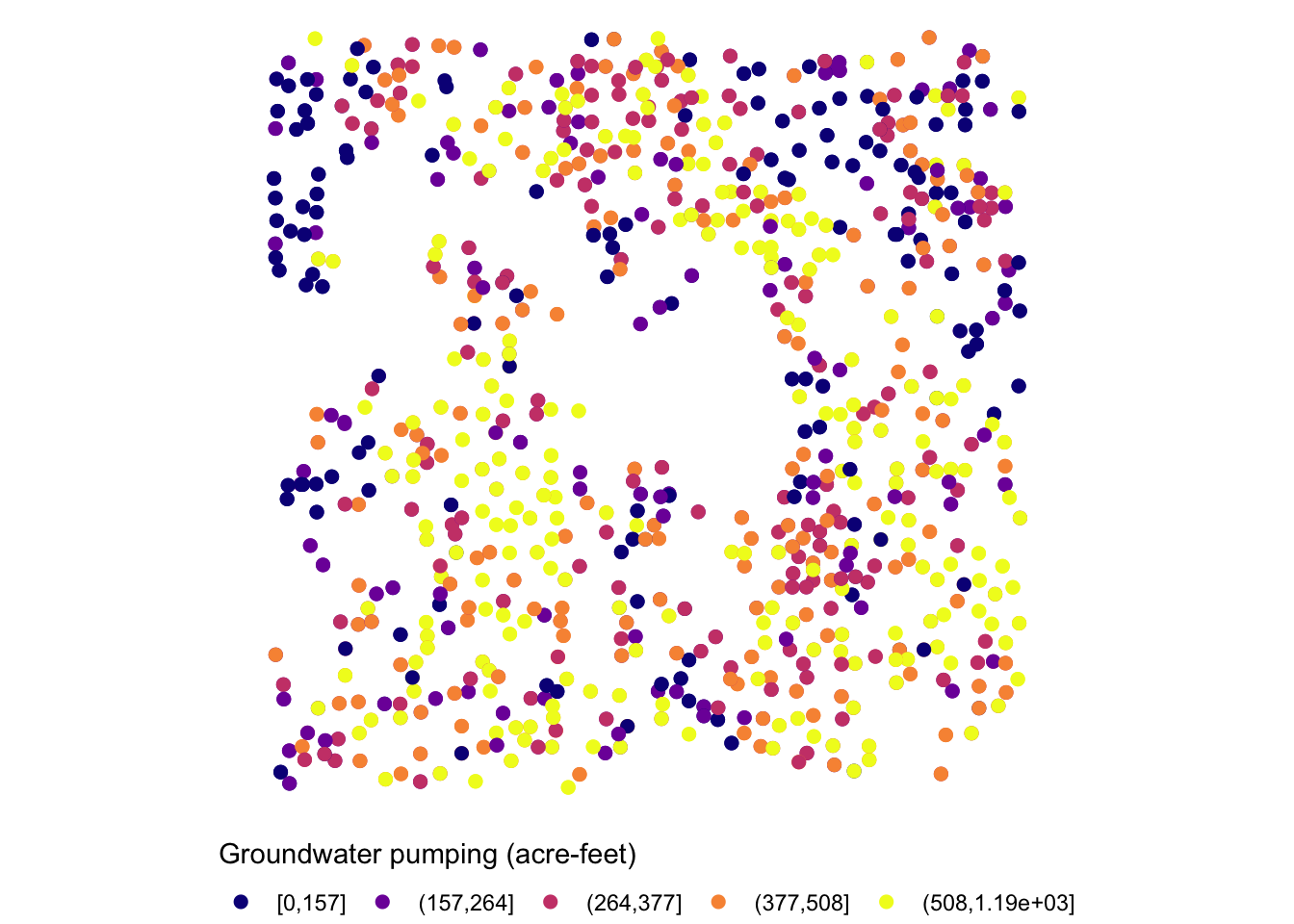
The legend is too long, so first put the legend title on top of the legend labels using the code below:
g_legend_2 +
guides(
color = guide_legend(title.position = "top")
)
Since the legend is for the color aesthetic attribute, color = guide_legend() was used. The legend labels are still a bit too long, so let’s arrange them in two rows using the nrow = option.
g_legend_2 +
guides(
color = guide_legend(title.position = "top", nrow = 2)
)
For the other options for guide_legend(), see here.
8.5.3 Facet strips
Facet strips refer to the area of boxed where the values of faceting variables are printed. In the figure below, it’s gray strips on top of the maps. You can change how they look using strip.* options in theme() and also partially inside facet_wrap() and facet_grid(). Here is the list of available options:
-
strip.background,strip.background.x,strip.background.y strip.placement-
strip.text,strip.text.x,strip.text.y strip.switch.pad.gridstrip.switch.pad.wrap
ggplot() +
#--- KS county boundary ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- wells ---#
geom_sf(data = gw_KS_sf, aes(color = af_used)) +
#--- facet by year (side by side) ---#
facet_wrap((af_used > 500) ~ year) +
theme_void() +
scale_color_viridis_c() +
theme(legend.position = "bottom")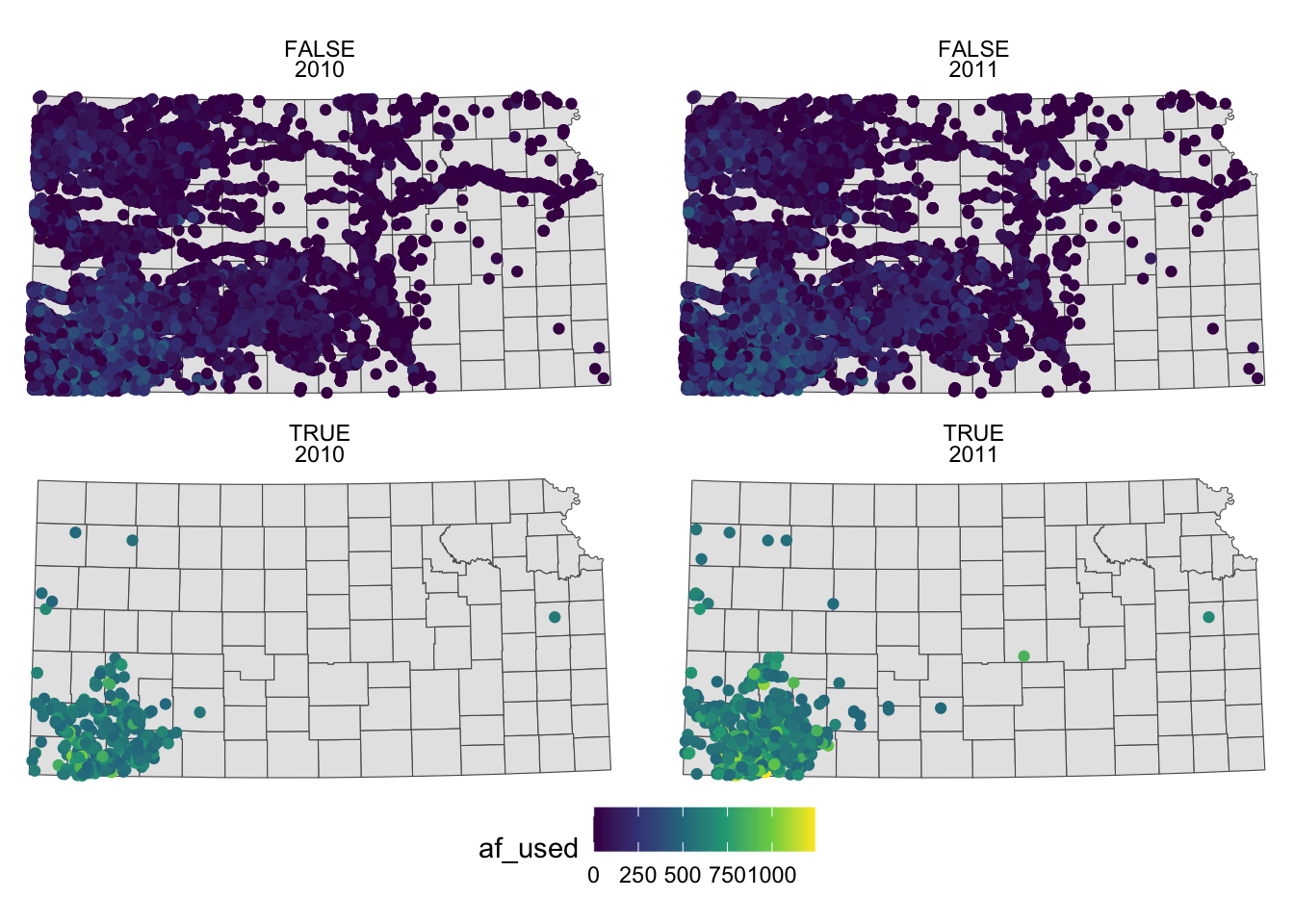
To make texts in the strips more descriptive of what they actually mean you can make variable that have texts you want to show on the map as their values.
gw_KS_sf <-
gw_KS_sf %>%
mutate(
high_low = ifelse(af_used > 500, "High water use", "Low water use"),
year_txt = paste("Year: ", year)
)
(
g_facet <-
ggplot() +
#--- KS county boundary ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- wells ---#
geom_sf(data = gw_KS_sf, aes(color = af_used)) +
#--- facet by year (side by side) ---#
facet_wrap(high_low ~ year_txt) +
theme_void() +
scale_color_viridis_c() +
theme(legend.position = "bottom")
)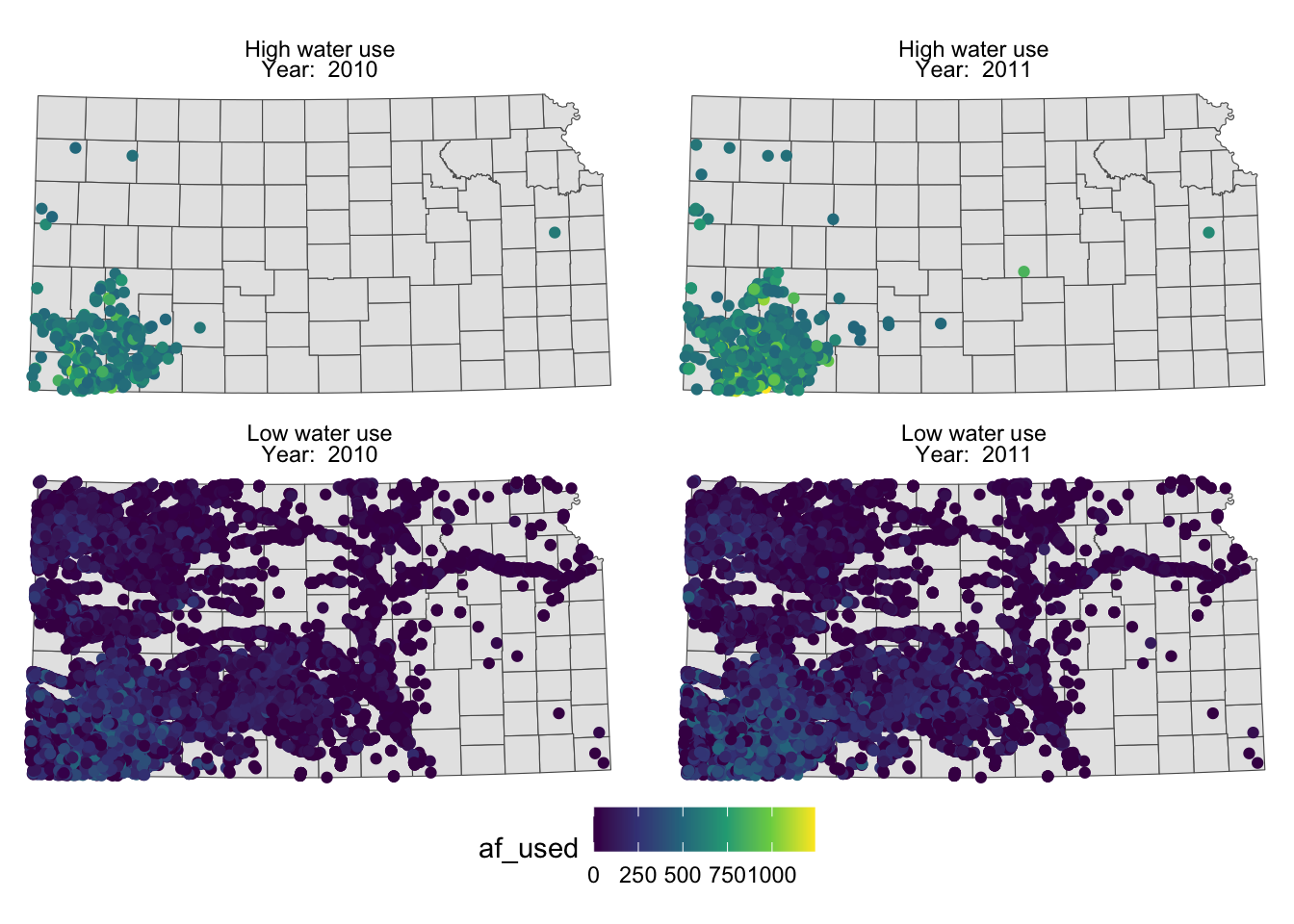
You probably noticed that the high water use cases are now appear on top. This is because the panels of figures are arranged in a way that the strip texts are alphabetically ordered. High water use precedes Low water use. Sometimes, this is not desirable. To force a specific order, you can turn the faceting variable (here high_low) into a factor with the order of its values defined using levels =. The following code converts high_low into a factor where “Low water use” is the first level and “High water use” is the second level.
gw_KS_sf <-
mutate(
gw_KS_sf,
high_low = factor(
high_low,
levels = c("Low water use", "High water use")
)
)Now, “Low water use” cases appear first (on top).
g_facet <-
ggplot() +
#--- KS county boundary ---#
geom_sf(data = st_transform(KS_county, 32614)) +
#--- wells ---#
geom_sf(data = gw_KS_sf, aes(color = af_used)) +
#--- facet by year (side by side) ---#
facet_wrap(high_low ~ year_txt) +
theme_void() +
scale_color_viridis_c() +
theme(legend.position = "bottom")
g_facet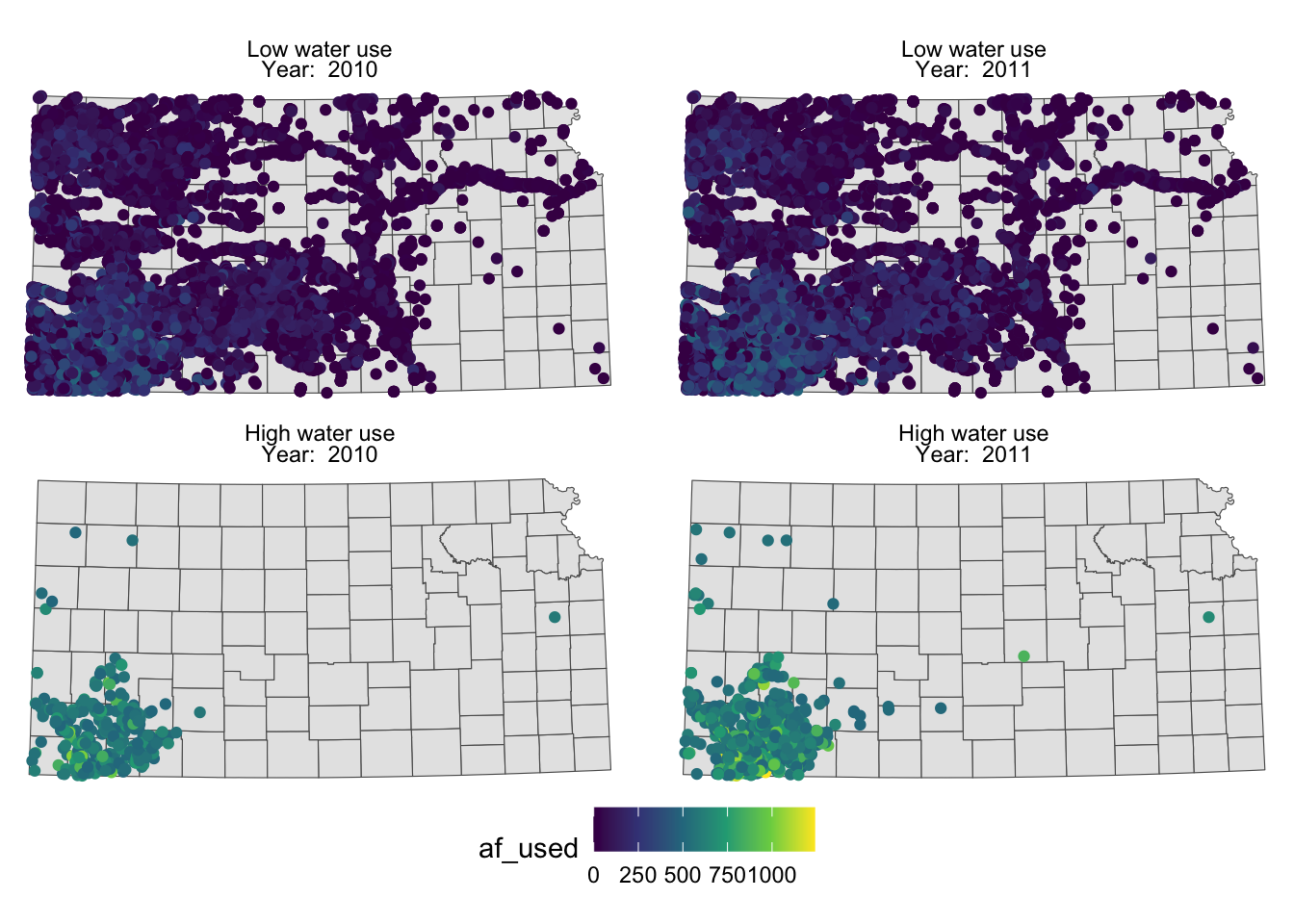
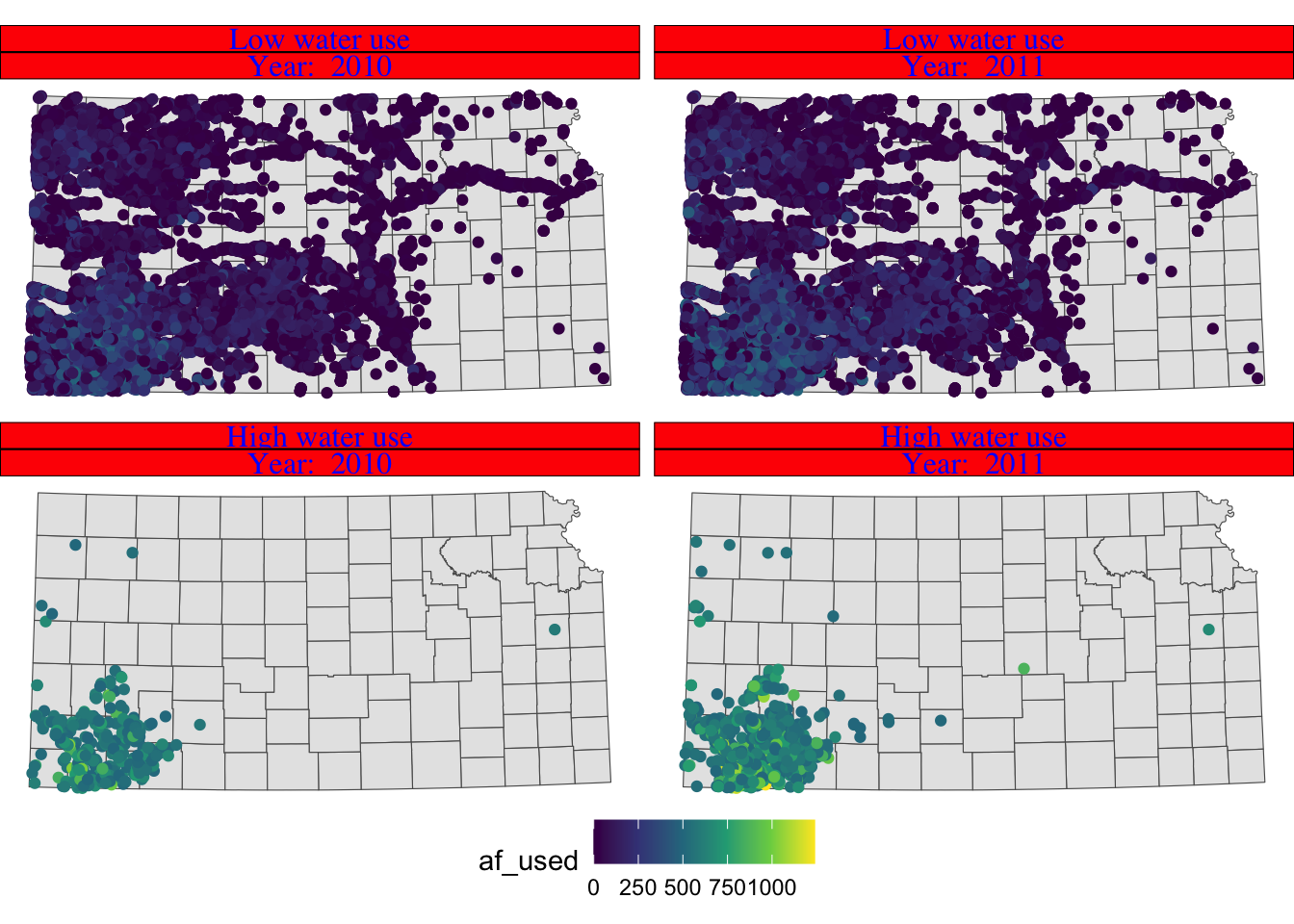
You can control how strip texts and strip boxes appear using strip.text and strip.background options. Here is an example:
g_facet + theme(
strip.text.x = element_text(size = 12, family = "Times", color = "blue"),
strip.background = element_rect(fill = "red", color = "black")
)
Instead of having descriptions of cases on top of the figures, you could have one of the descriptions on the right side of the figures using facet_grid().
g_facet +
#--- this overrides facet_wrap(high_low ~ year_txt) ---#
facet_grid(high_low ~ year_txt)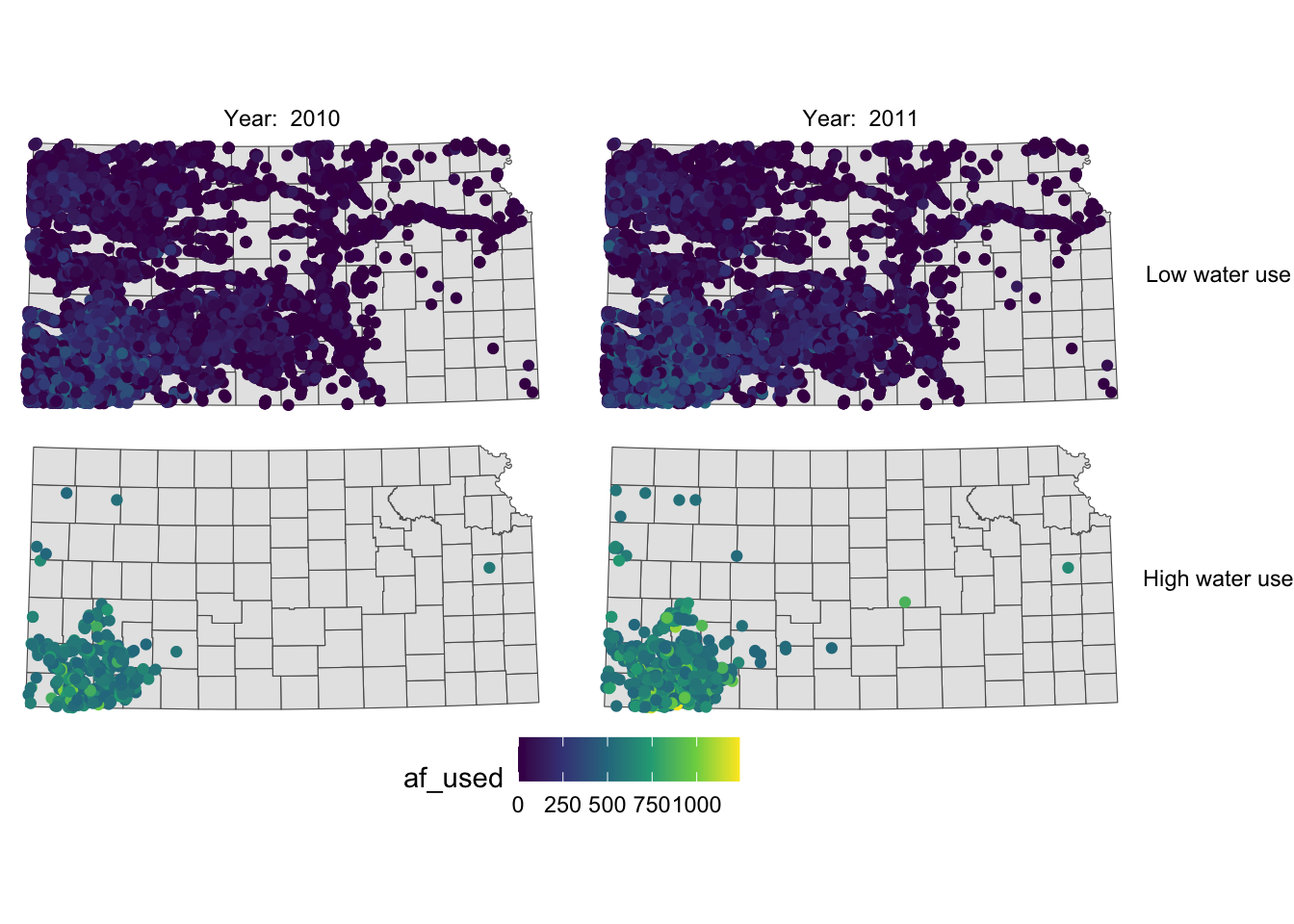
Now case descriptions for high_low are too long and it is squeezing the space for maps. Let’s flip high_low and year.
g_facet +
#--- this overrides facet_grid(high_low ~ year_txt) ---#
facet_grid(year_txt ~ high_low)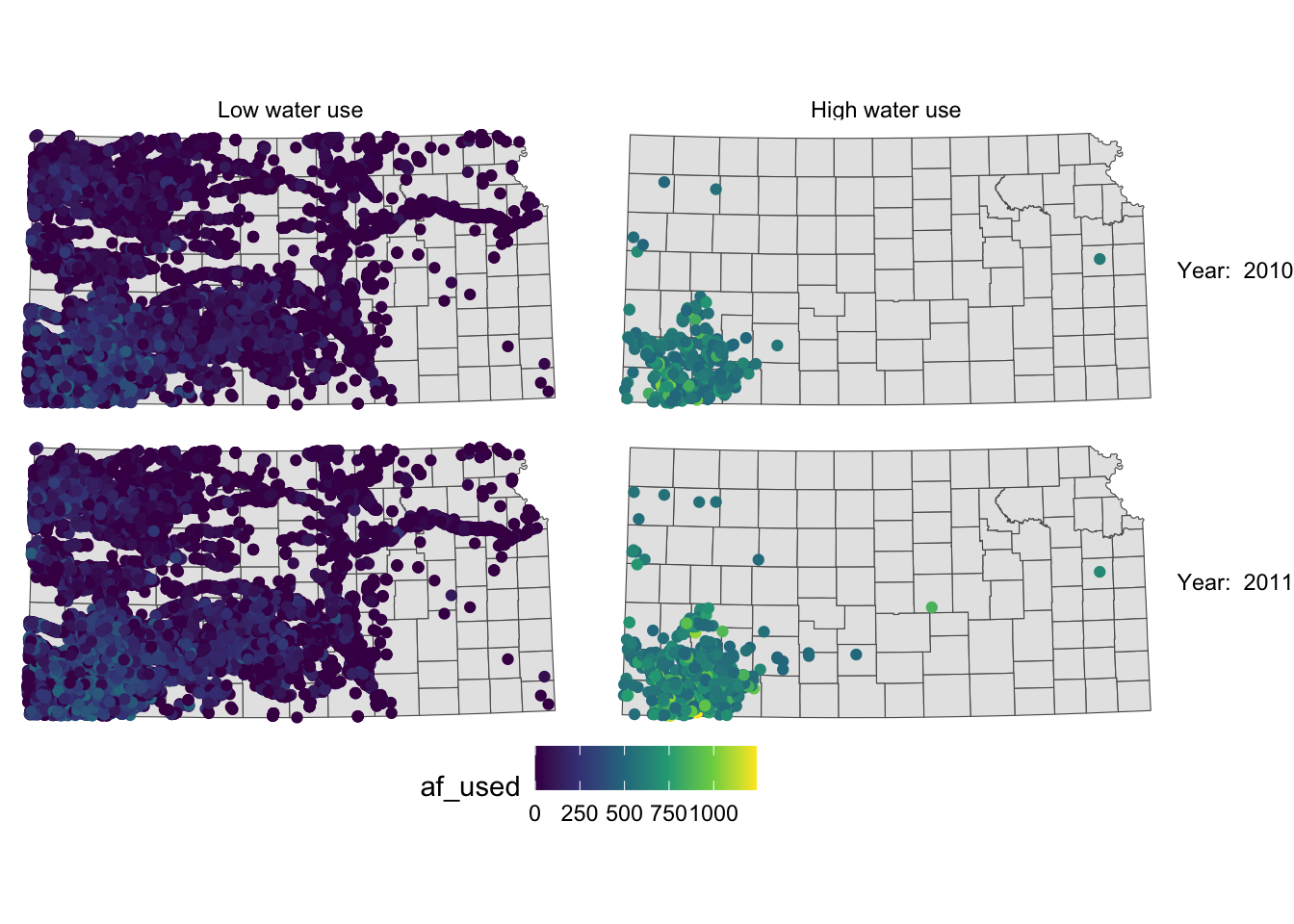
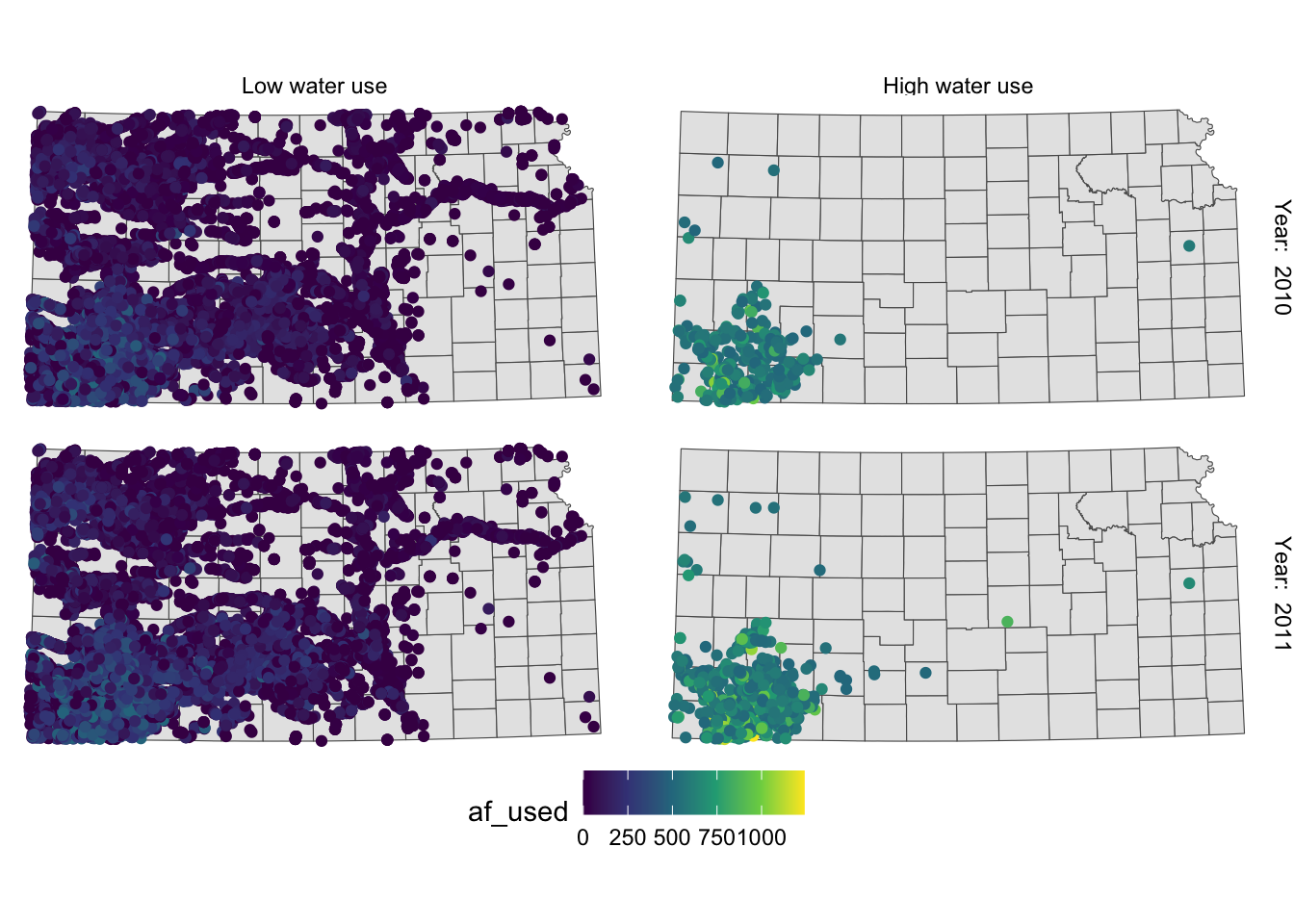
This is slightly better than before, but not much. Let’s rotate strip texts for year using the angle = option.
g_facet +
#--- this overrides facet_grid(high_low ~ year_txt) ---#
facet_grid(year_txt ~ high_low) +
theme(
strip.text.y = element_text(angle = -90)
)
Since we only want to change the angle of strip texts for the second faceting variable, we need to work on strip.text.y (if you want to work on the first one, you use strip.text.x.).
Let’s change the size of the strip texts to 12 and use Times font family.
g_facet +
#--- this overrides facet_grid(high_low ~ year_txt) ---#
facet_grid(year_txt ~ high_low) +
theme(
strip.text.y = element_text(angle = -90, size = 12, family = "Times"),
#--- moves up the strip texts ---#
strip.text.x = element_text(size = 12, family = "Times")
)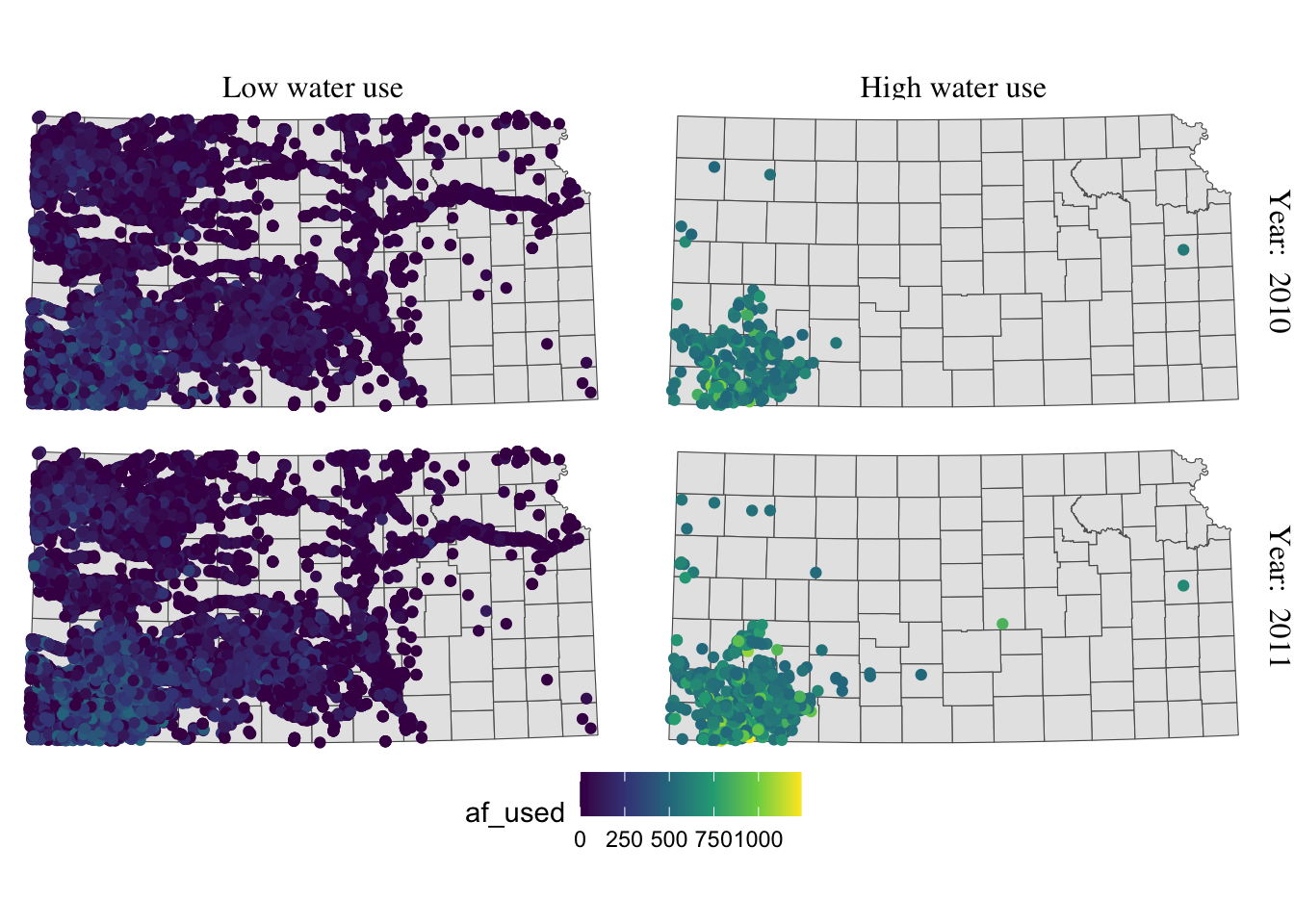
The strip texts for high_low are too close to maps that letter “g” in “High” is truncated. Let’s move them up.
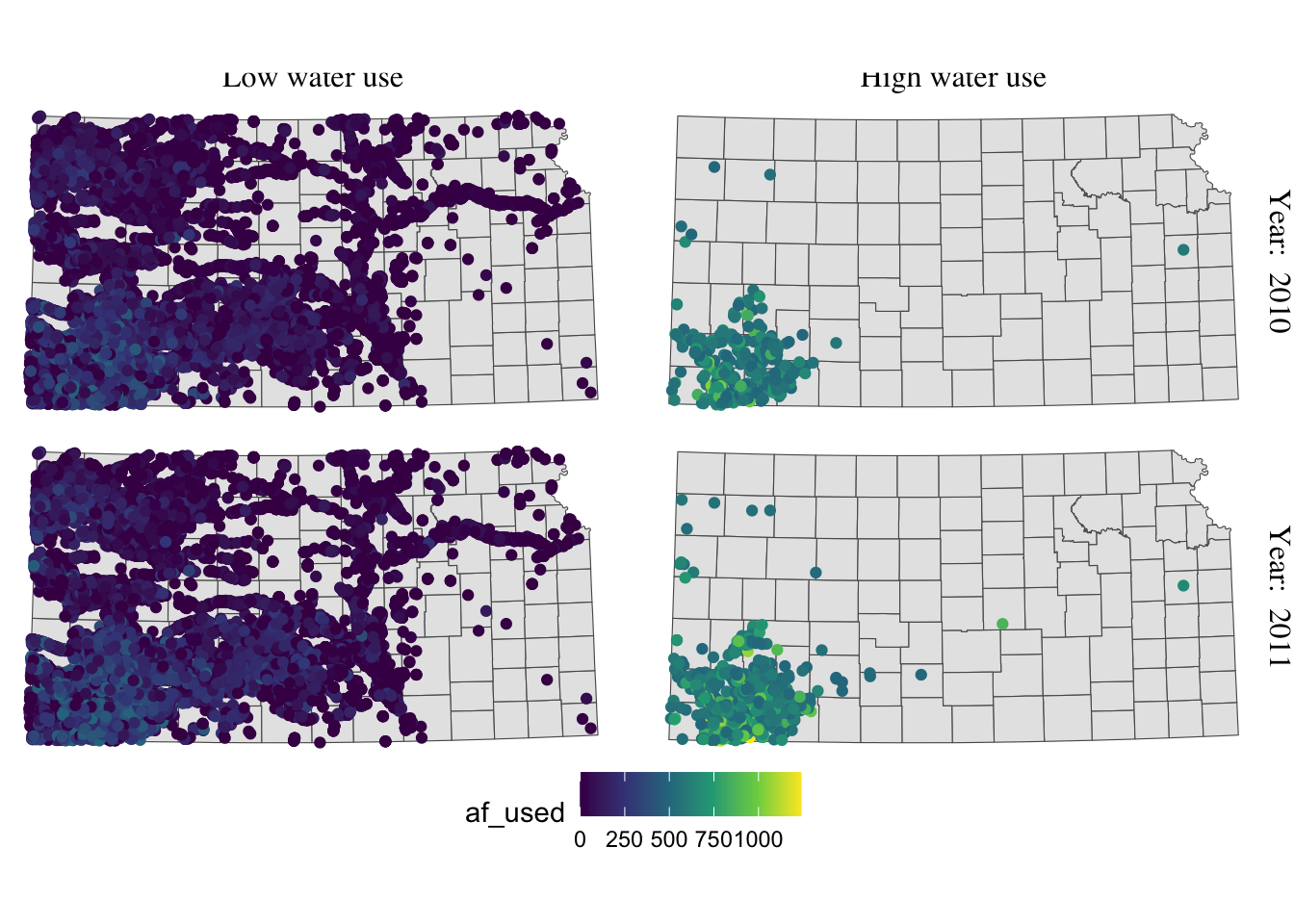
g_facet +
#--- this overrides facet_grid(high_low ~ year_txt) ---#
facet_grid(year_txt ~ high_low) +
theme(
strip.text.y = element_text(angle = -90, size = 12, family = "Times"),
#--- moves up the strip texts ---#
strip.text.x = element_text(vjust = 2, size = 12, family = "Times")
)
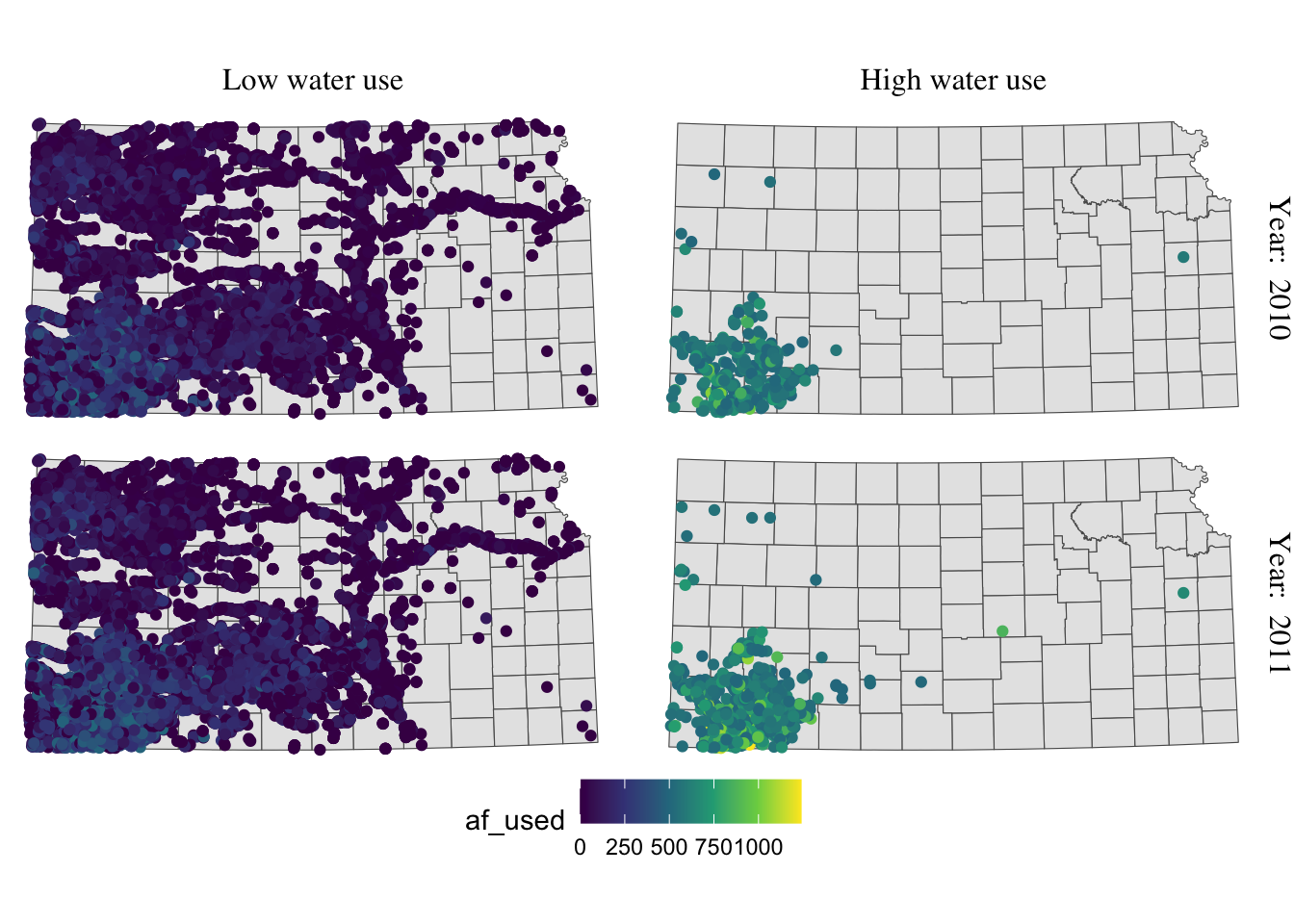
Now the upper part of the letters is truncated. We could just put more margin below the texts using the margin = margin(top, right, bottom, left, unit in text) option.
g_facet +
#--- this overrides facet_grid(high_low ~ year_txt) ---#
facet_grid(year_txt ~ high_low) +
theme(
strip.text.y = element_text(angle = -90, size = 12, family = "Times"),
strip.text.x = element_text(margin = margin(0, 0, 0.2, 0, "cm"), size = 12, family = "Times")
)
For completeness, let’s make the legend look better as well (this is discussed in section 8.5.1).
(
g_facet <- g_facet +
#--- this overrides facet_grid(high_low ~ year_txt) ---#
facet_grid(year_txt ~ high_low) +
theme(
strip.text.y = element_text(angle = -90, size = 12, family = "Times"),
strip.text.x = element_text(margin = margin(0, 0, 0.2, 0, "cm"), size = 12, family = "Times")
) +
theme(legend.position = "bottom") +
labs(color = "Groundwater pumping (acre-feet)") +
theme(
legend.position = "bottom",
legend.key.height = unit(0.5, "cm"),
legend.key.width = unit(2, "cm"),
legend.text = element_text(size = 12, family = "Times"),
legend.title = element_text(size = 12, family = "Times")
#--- NEW LINES HERE!! ---#
) +
guides(color = guide_colorbar(title.position = "top"))
)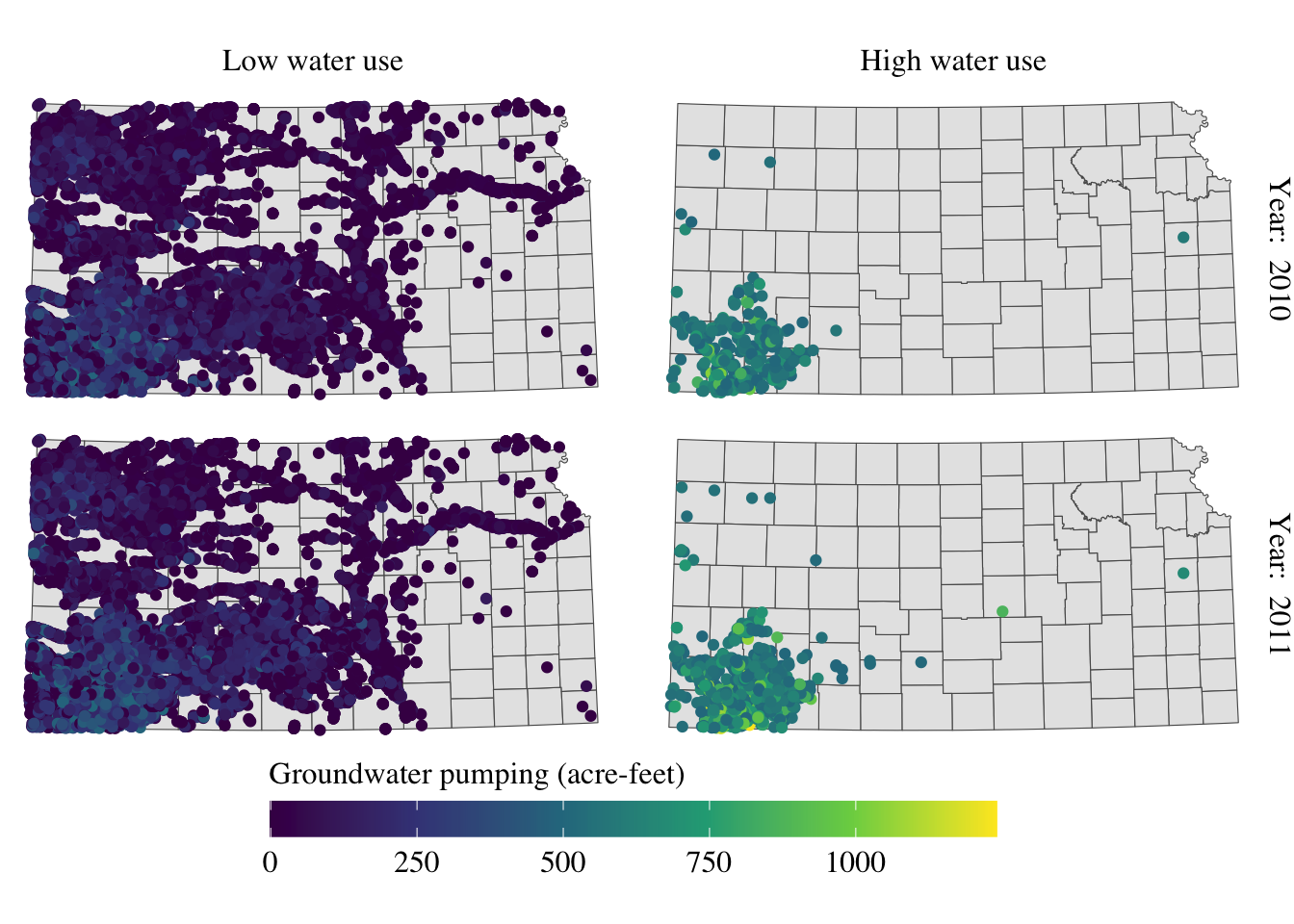
Alright, setting aside the problem of whether the information provided in the maps is meaningful or not, the maps look great at least.
8.5.4 North arrow and scale bar
The ggspatial package lets you put a north arrow and scale bar on a map using annotation_scale() and annotation_north_arrow().
#--- get North Carolina county borders ---#
nc <- st_read(system.file("shape/nc.shp", package = "sf"))Reading layer `nc' from data source
`/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/sf/shape/nc.shp'
using driver `ESRI Shapefile'
Simple feature collection with 100 features and 14 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
Geodetic CRS: NAD27
(
#--- create a simple map ---#
g_nc <- ggplot(nc) +
geom_sf() +
theme_void()
)
Here is an example code that adds a scale bar:
#--- load ggspatial ---#
library(ggspatial)
#--- add scale bar ---#
g_nc +
annotation_scale(
location = "bl",
width_hint = 0.2
)location determines where the scale bar is. The first letter is either t (top) or b (bottom), and the second letter is either l (left) or r (right). width_hint is the length of the scale bar relative to the plot. The distance number (200 km) was generated automatically according to the length of the bar.
You can add pads from the plot border to fine tune the location of the scale bar:
g_nc +
annotation_scale(
location = "bl",
width_hint = 0.2,
pad_x = unit(3, "cm"),
pad_y = unit(2, "cm")
)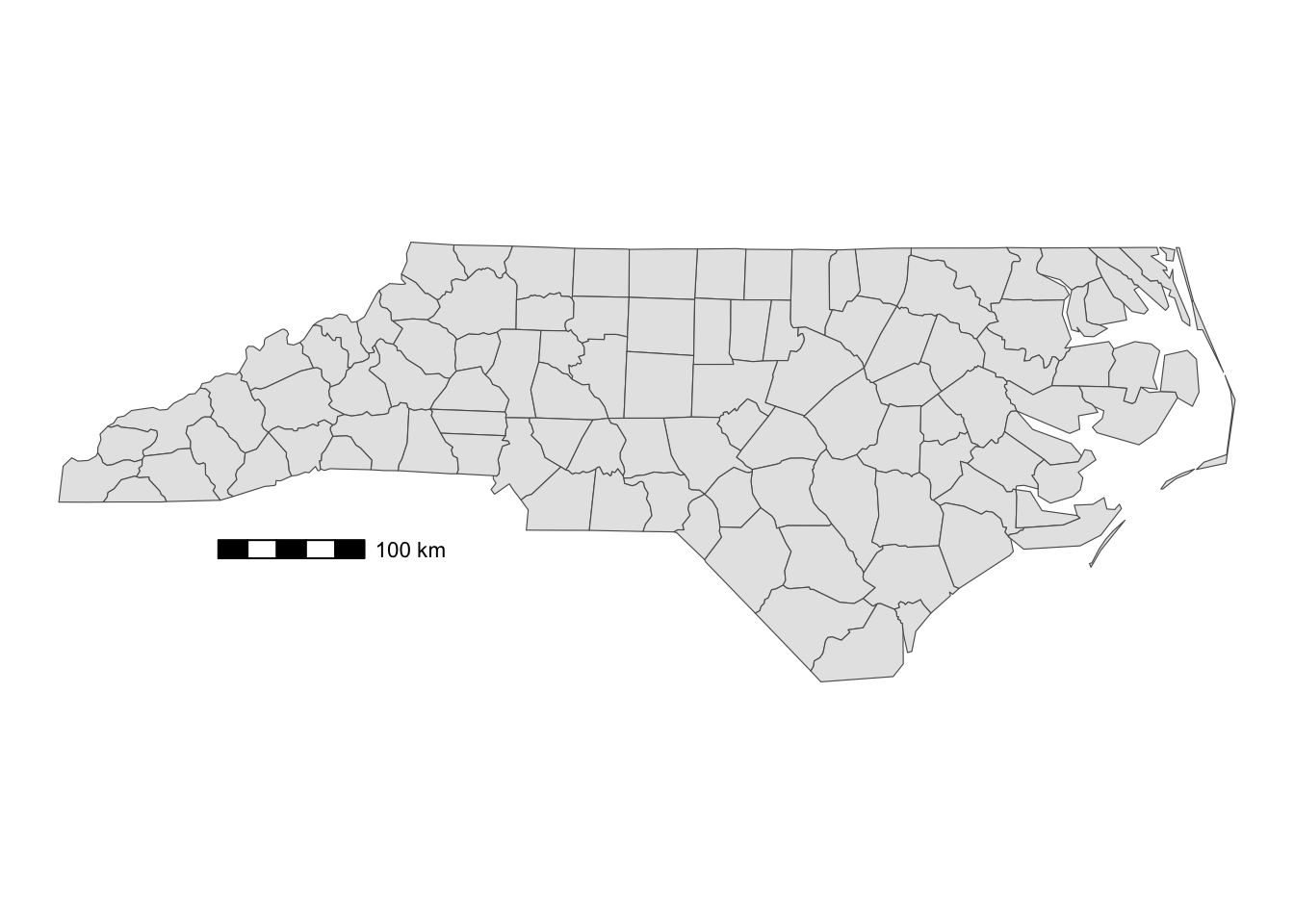
A positive number means that the scale bar will be placed further away from closest border of the plot.
You can add a north arrow using annotation_north_arrow(). Accepted arguments are similar to those for annotation_scale().
g_nc +
annotation_scale(
location = "bl",
width_hint = 0.2,
pad_x = unit(3, "cm")
) +
#--- add north arrow ---#
annotation_north_arrow(
location = "tl",
pad_x = unit(0.5, "in"),
pad_y = unit(0.1, "in"),
style = north_arrow_fancy_orienteering
)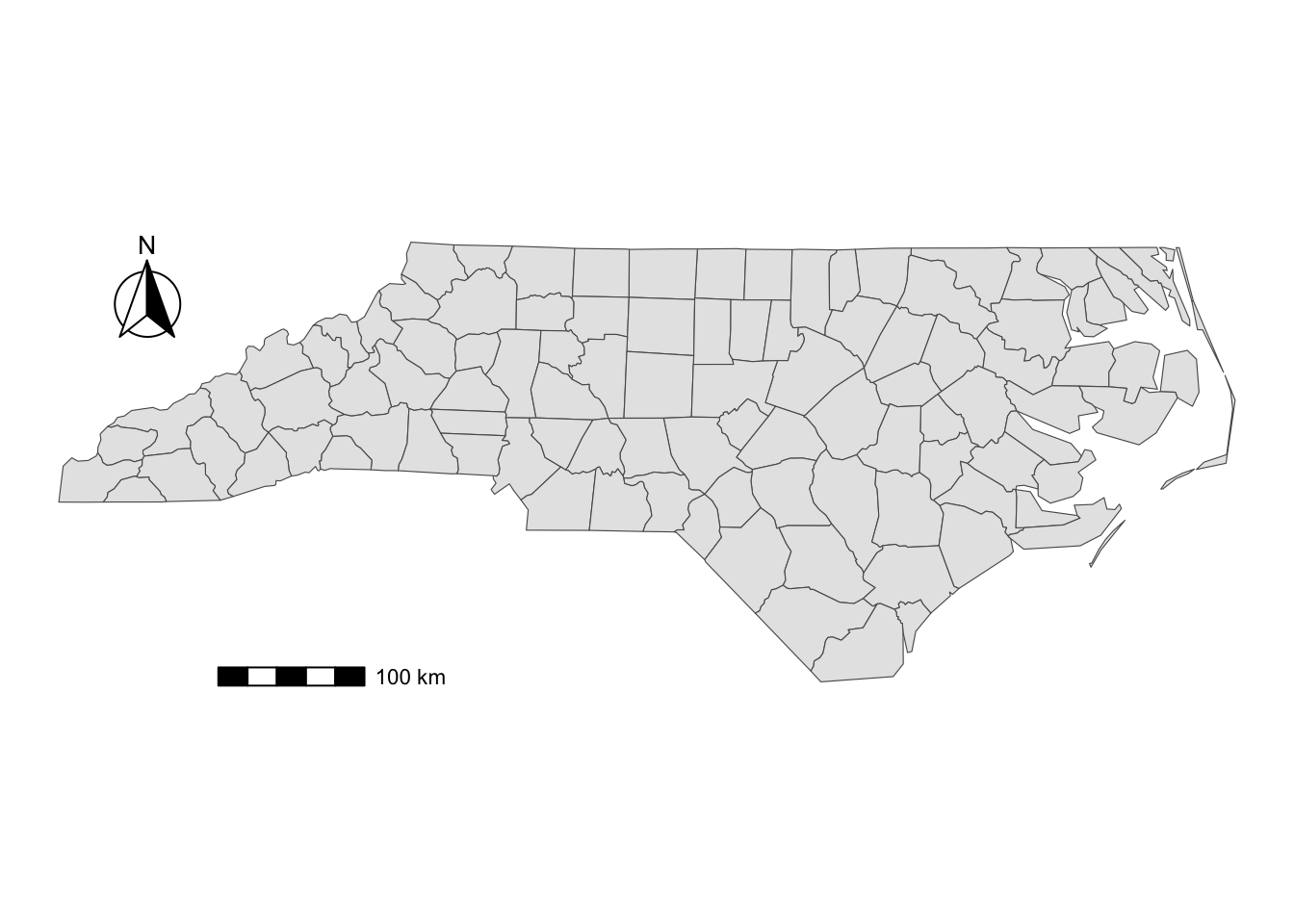
There are several styles you can pick from. Run ?north_arrow_orienteering to see other options.
8.6 Saving a ggplot object as an image
Maps created with ggplot2 can be saved using ggsave() with the following syntax:
ggsave(filename = file name, plot = ggplot object)
#--- or just this ---#
ggsave(file name, ggplot object)Many different file formats are supported including pdf, svg, eps, png, jpg, tif, etc. One thing you want to keep in mind is the type of graphics:
- vector graphics (pdf, svg, eps)
- raster graphics (jpg, png, tif)
While vector graphics are scalable, raster graphics are not. If you enlarge raster graphics, the cells making up the figure become visible, making the figure unappealing. So, unless it is required to save figures as raster graphics, it is encouraged to save figures as vector graphics.94
Let’s try to save the following ggplot object.
#--- get North Carolina county borders ---#
nc <- st_read(system.file("shape/nc.shp", package = "sf"))Reading layer `nc' from data source
`/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/sf/shape/nc.shp'
using driver `ESRI Shapefile'
Simple feature collection with 100 features and 14 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
Geodetic CRS: NAD27
ggsave() automatically detects the file format from the file name. For example, the following code saves g_nc as nc.pdf. ggsave() knows the ggplot object was intended to be saved as a pdf file from the extension of the specified file name.
ggsave("nc.pdf", g_nc)Similarly,
#--- save as an eps file ---#
ggsave("nc.eps", g_nc)
#--- save as an eps file ---#
ggsave("nc.svg", g_nc)You can change the output size with height and width options. For example, the following code creates a pdf file of height = 5 inches and width = 7 inches.
ggsave("nc.pdf", g_nc, height = 5, width = 7)You change the unit with the units option. By default, in (inches) is used.
You can control the resolution of the output image by specifying DPI (dots per inch) using the dpi option. The default DPI value is 300, but you can specify any value suitable for the output image, including “retina” (320) or “screen” (72). 600 or higher is recommended when a high resolution output is required.