06-1: How to write R codes, manage Projects, work with RStudio
Tips to make the most of the lecture notes
Click on the three horizontally stacked lines at the bottom left corner of the slide, then you will see table of contents, and you can jump to the section you want
Hit letter “o” on your keyboard and you will have a panel view of all the slides
- The box area with a hint of blue as the background color is where you can write code (hereafter referred to as the “code area”).
- Hit the “Run Code” button to execute all the code inside the code area.
- You can evaluate (run) code selectively by highlighting the parts you want to run and hitting Command + Enter for Mac (Ctrl + Enter for Windows).
- If you want to run the codes on your computer, you can first click on the icon with two sheets of paper stacked on top of each other (top right corner of the code chunk), which copies the code in the code area. You can then paste it onto your computer.
- You can click on the reload button (top right corner of the code chunk, left to the copy button) to revert back to the original code.
Learning objectives
- Learn how to organize your project: folder, codes, data files, etc
- Learn how to organize R codes
- Learn how to use various RStudio tips for efficient programming
Reproducibility
Reproducibility
First of all, you may have heard of “reproducibility” and “replicability.” While they sometimes are used interchangeably, they mean different things. Here are commonly used definitions of the two terms (Cacioppo et al. 2015).
Definition: Reproducibility
A research study is reproducible if anybody (including the author of the study) can generate exactly the same results by using the same materials (e.g., data) and procedures used in the study.
Definition: Replicability
A research study is replicable if other teams reach the same conclusion by applying the same procedure to the different materials (e.g., data).
This lecture focuses only on reproducibility and do not deal with replicability.
Minimum Requirement
Every single action taken during the entire research process is documented in a way that anybody can follow to implement the same actions (no hidden actions) to produce exactly the same results.
Note that this does not necessarily mean every single action needs to be computer-programmed and automated. Even if you manually delete rows of data on Excel (highly discouraged), this does not make your research non-reproducible as long as this action is recorded and the original data (before deletion of the rows) are provided because anybody can implement this action.
A project can be considered reproducible if the minimum requirements are met. However, it is still low-quality if reproducing it requires excessive time or resources. A high-quality reproducible project, on the other hand, demonstrates the following characteristics:
Organized Project Folder
A well-structured folder system makes it easy to locate files and navigate the project.
Streamlined Automation
Workflows are automated through clearly annotated programs, making replication straightforward and ensuring transparency in the process.
Comprehensive Documentation
Thorough documentation—including details on data, methods, and reproduction steps—saves time and enhances clarity for anyone reproducing the work.
The main beneficiaries of reproducible research include:
- You (Y)
- Members of your team (M)
- The scientific community (S)
Here are some the benefits of high-quality reproducible research with their beneficiaries:
- Scientific Integrity (S)
- Educational Value (MS)
- Repeatability (YM)
- Transferability (YM)
- Reducing Errors (YM)
How to organize your project
How to organize your project
You should have a single dedicated folder for a research project. This will avoid
confusions between objects of the same or similar name (accidentally using the one you do not intend to use)
wasting memory by holding objects on the global environment that are completely irrelevant to your working project
We can initiate an RStudio project with a dedicated folder from within RStudio.
At the top right corner of RStudio, navigate through:
Project (None)
-> New Project…
-> New Directory
-> New Project
- type in a directory name
- select the directory in which the project folder (directory) is going to be created
- hit the create folder button
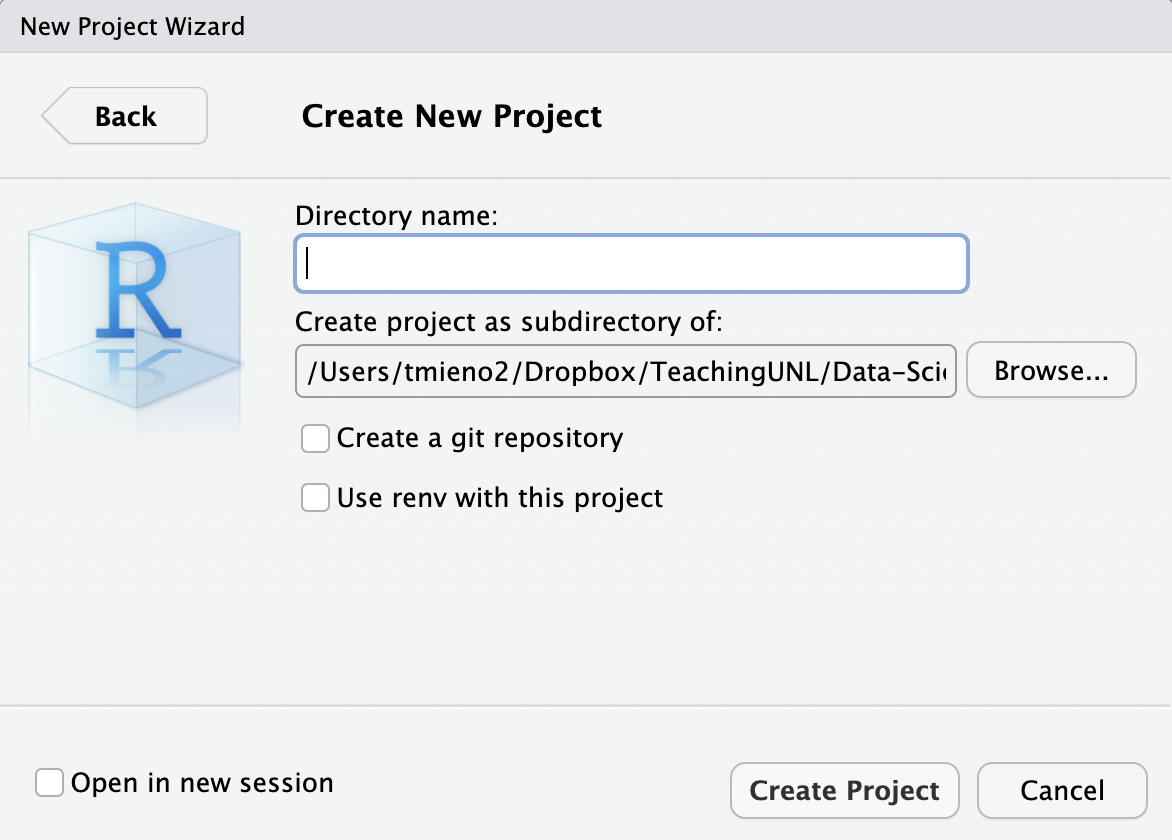
You will be automatically taken to a new R session inside the newly-created RStudio project.
In this folder you just created, you have a single file named
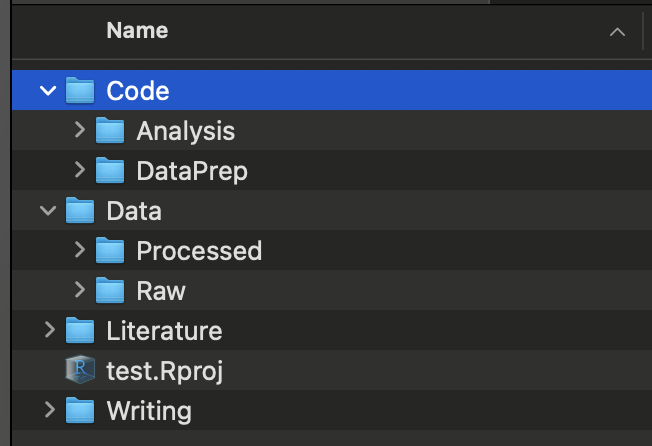
Here is a recommended folder organization. You should modify/add folders as you see fit.
- Code: all the codes go in here
- DataPrep:
- Analysis:
- Data
- Raw: place raw datasets here
- Processed: save intermediate datasets here
- Literature: journal articles and other relevant documents
- Results: results (regression, figures, tables)
- Writing: qmd, WORD, Latex files
Files
Recommendation
Use a qmd file instead of an R file whenever you write codes
It is much easier to make comments in a qmd file than an R file
You can better organize your codes with markdown section headers (e.g., #, ##)
R crashed at a certain chunk and had to restart R and then run all the R codes up to the problematic chunk? Use
Run All Chunks Abovebutton (click on the triangle right to theRunbutton and select the option, or hitoption+command+P).Easily move between sections and subsection using the navigator at the bottom lower corner of the source pane
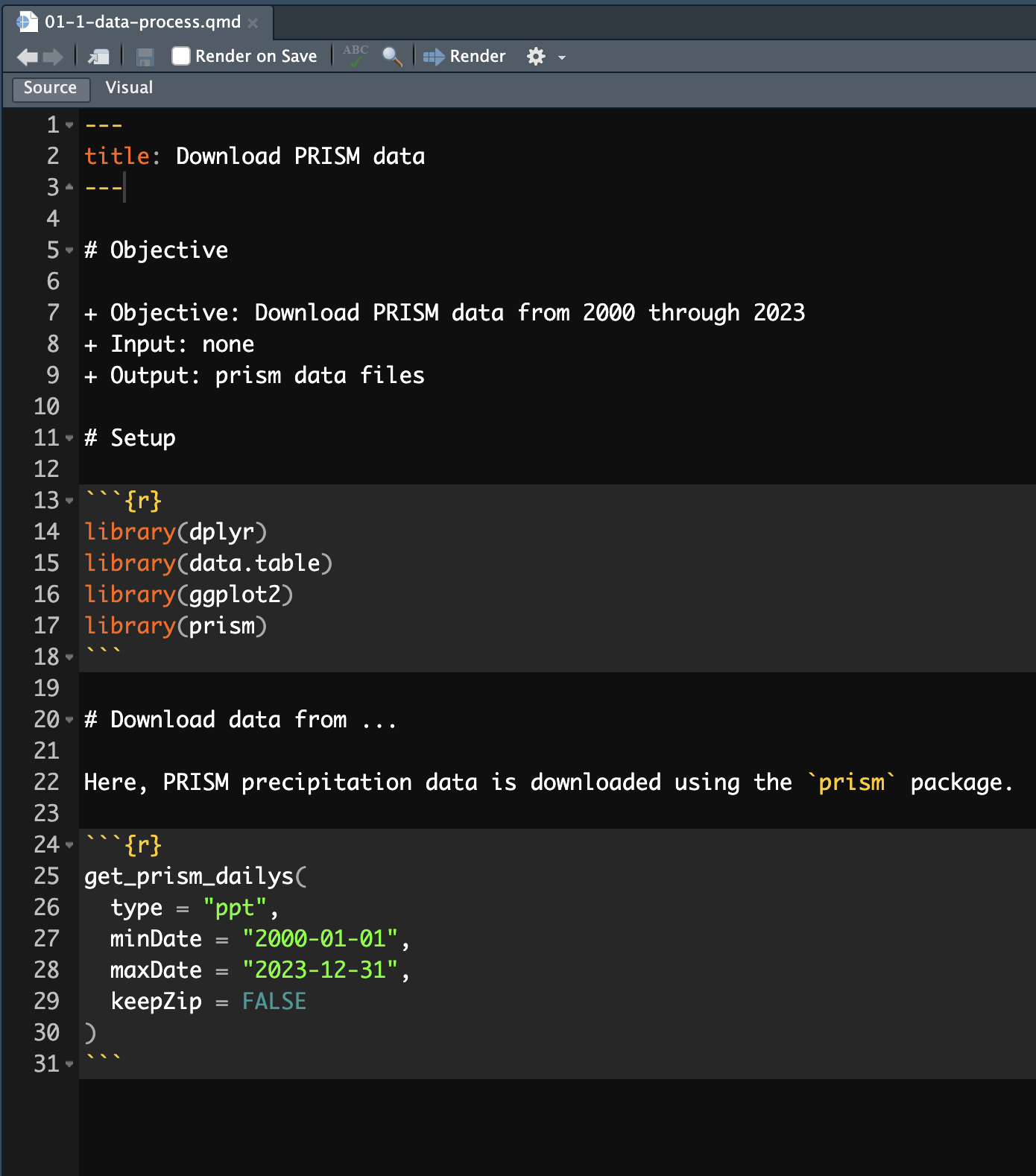
Recommended Stucture
- Objective statement
- state objectives
- input: state input files and datasets
- output: state output files and datasets
- Setup
- set the working directory (if necessary)
- load packages
- load functions
- Actions
- Action 1
- Action 2
- .
- .
- .
- Action n
Note
Dynamically edit the “Objective statement” as its objectives, input, and output are likely to change.
You can write R codes however you would like. But, your code may get more readable to you and others who might read your codes by following a style guideline that is accepted by many R users. There are several popular styles of formatting R codes:
Examples
Here are some examples of the tidyverse style:
The styler package can help you follow partially the tidyverse coding style.
Once the package is installed, you can highlight the lines of codes and hit cmd + shift + A for Mac (ctrl + shift + A for Windows) to reformat the codes to conform with the tidyverse style.
Alternatively, you can click on Addins in the middle of the menu at the top, and select style selection.
Rules 1
- place all the raw datasets (nothing else) in a designated folder inside the Data/Raw folder
- do not ever override them, you only read them and keep them intact
Rules 2
- write R codes to process (transform, merge, etc) the raw data and save all the R codes inside the Code/DataPrep folder
- save intermediate R objects or datasets in the Data/Processed folder
- do not mix codes and datasets in a single folder
Rules 3
- write R codes to do analysis and save them in Code/Analysis
- save the results/outputs (regression tables, figures, tables) in the Results folder
Rules 4
- if you are using qmd to write a journal article or report, put them in the Writing folder (same goes for WORD)
- refer to figures and tables in the Results folder to integrate them in the output document
Recommendation
- Name files so that you know what purposes they serve for you
- Place numbers as prefix to indicate the order in which they should be run
Example
- Data Collection and Preparation (in Code/DataPrep)
- 01-1-download-weather-data.R
- 01-2-download-political-boundary-data.R
- 01-3-summarize-data.R
- 01-4-merge-data.R
- Data Analysis and Results Preparation (in Code/Analysis)
- 02-1-regression-analysis.R
- 02-2-gen-figures-tables.R
Example project
Let’s take a look at an example project that is designed to be reproducible. First, Clone this repository. We will then look at how the project is organized and developed.
RStudio tips
Code snippets
Code snippets are functions that maps sequence of letters and symbols (short) to other sequence of letters and symbols (more complicated and long)
Syntax
Important
You need a tab before (what you want to print)
Follow Tools \(\rightarrow\) Global Options \(\rightarrow\) Code \(\rightarrow\) Edit Snippets , select R tab, and add snippets.
Suppose you are working on an Quarto document.
- You are in the R context if your cursor is in an R code chunk
- You are in the Markdown context if your cursor is outside of an R code chunk
Snippets defined in the R (Markdown) tab only works in the R (Markdown) context.
$ is used as a special character to denote where the cursor should jump (by hitting tab) after completing each section of a snippet.